- Record: found
- Abstract: found
- Article: found
Thiazoline-related innate fear stimuli orchestrate hypothermia and anti-hypoxia via sensory TRPA1 activation
Read this article at
Abstract
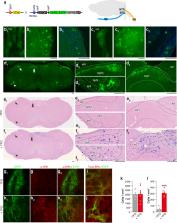
Thiazoline-related innate fear-eliciting compounds (tFOs) orchestrate hypothermia, hypometabolism, and anti-hypoxia, which enable survival in lethal hypoxic conditions. Here, we show that most of these effects are severely attenuated in transient receptor potential ankyrin 1 ( Trpa1) knockout mice. TFO-induced hypothermia involves the Trpa1-mediated trigeminal/vagal pathways and non- Trpa1 olfactory pathway. TFOs activate Trpa1-positive sensory pathways projecting from trigeminal and vagal ganglia to the spinal trigeminal nucleus (Sp5) and nucleus of the solitary tract (NTS), and their artificial activation induces hypothermia. TFO presentation activates the NTS-Parabrachial nucleus pathway to induce hypothermia and hypometabolism; this activation was suppressed in Trpa1 knockout mice. TRPA1 activation is insufficient to trigger tFO-mediated anti-hypoxic effects; Sp5/NTS activation is also necessary. Accordingly, we find a novel molecule that enables mice to survive in a lethal hypoxic condition ten times longer than known tFOs. Combinations of appropriate tFOs and TRPA1 command intrinsic physiological responses relevant to survival fate.
Abstract
Matsuo et al. report that thiazoline-related innate fear-eliciting compounds activate the spinal trigeminal nucleus (Sp5) and the nucleus of the solitary tract (NTS) via vagal/trigeminal TRPA1 to induce robust physiological alterations, enabling long time survival in a lethal hypoxic environment.
Related collections
Most cited references59

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
Near-optimal probabilistic RNA-seq quantification.
- Record: found
- Abstract: found
- Article: not found