- Record: found
- Abstract: found
- Article: found
Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease
Read this article at
Abstract
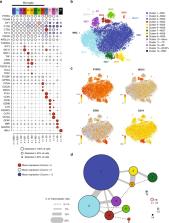
The extent of microglial heterogeneity in humans remains a central yet poorly explored question in light of the development of therapies targeting this cell type. Here, we investigate the population structure of live microglia purified from human cerebral cortex samples obtained at autopsy and during neurosurgical procedures. Using single cell RNA sequencing, we find that some subsets are enriched for disease-related genes and RNA signatures. We confirm the presence of four of these microglial subpopulations histologically and illustrate the utility of our data by characterizing further microglial cluster 7, enriched for genes depleted in the cortex of individuals with Alzheimer’s disease (AD). Histologically, these cluster 7 microglia are reduced in frequency in AD tissue, and we validate this observation in an independent set of single nucleus data. Thus, our live human microglia identify a range of subtypes, and we prioritize one of these as being altered in AD.
Abstract
Imbalance of microglial phenotypes in the aging brain might underlie their involvement in late onset neurodegenerative diseases. Here we report the population structure of microglia in the aged human brain and the reduction of a particular microglia subset in individuals with Alzheimer’s disease .
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
edgeR: a Bioconductor package for differential expression analysis of digital gene expression data
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found