- Record: found
- Abstract: found
- Article: found
Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease
Read this article at
Abstract
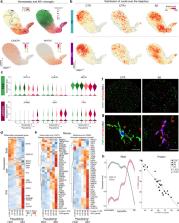
Alzheimer’s disease (AD) is the most prevalent form of dementia and is characterized by abnormal extracellular aggregates of amyloid-β and intraneuronal hyperphosphorylated tau tangles and neuropil threads. Microglia, the tissue-resident macrophages of the central nervous system (CNS), are important for CNS homeostasis and implicated in AD pathology. In amyloid mouse models, a phagocytic/activated microglia phenotype has been identified. How increasing levels of amyloid-β and tau pathology affect human microglia transcriptional profiles is unknown. Here, we performed snRNAseq on 482,472 nuclei from non-demented control brains and AD brains containing only amyloid-β plaques or both amyloid-β plaques and tau pathology. Within the microglia population, distinct expression profiles were identified of which two were AD pathology-associated. The phagocytic/activated AD1-microglia population abundance strongly correlated with tissue amyloid-β load and localized to amyloid-β plaques. The AD2-microglia abundance strongly correlated with tissue phospho-tau load and these microglia were more abundant in samples with overt tau pathology. This full characterization of human disease-associated microglia phenotypes provides new insights in the pathophysiological role of microglia in AD and offers new targets for microglia-state-specific therapeutic strategies.
Related collections
Most cited references35

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
edgeR: a Bioconductor package for differential expression analysis of digital gene expression data
- Record: found
- Abstract: found
- Article: not found