- Record: found
- Abstract: found
- Article: found
Single-Cell Analyses Reveal Megakaryocyte-Biased Hematopoiesis in Myelofibrosis and Identify Mutant Clone-Specific Targets
Read this article at
Summary
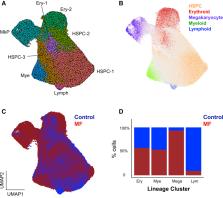
Myelofibrosis is a severe myeloproliferative neoplasm characterized by increased numbers of abnormal bone marrow megakaryocytes that induce fibrosis, destroying the hematopoietic microenvironment. To determine the cellular and molecular basis for aberrant megakaryopoiesis in myelofibrosis, we performed single-cell transcriptome profiling of 135,929 CD34 + lineage − hematopoietic stem and progenitor cells (HSPCs), single-cell proteomics, genomics, and functional assays. We identified a bias toward megakaryocyte differentiation apparent from early multipotent stem cells in myelofibrosis and associated aberrant molecular signatures. A sub-fraction of myelofibrosis megakaryocyte progenitors (MkPs) are transcriptionally similar to healthy-donor MkPs, but the majority are disease specific, with distinct populations expressing fibrosis- and proliferation-associated genes. Mutant-clone HSPCs have increased expression of megakaryocyte-associated genes compared to wild-type HSPCs, and we provide early validation of G6B as a potential immunotherapy target. Our study paves the way for selective targeting of the myelofibrosis clone and illustrates the power of single-cell multi-omics to discover tumor-specific therapeutic targets and mediators of tissue fibrosis.
Graphical Abstract
Highlights
-
•
Single-cell-omics demonstrate megakaryocyte-biased hematopoiesis in myelofibrosis (MF)
-
•
Megakaryocyte progenitors (MkPs) show high expression of a fibrosis signature in MF
-
•
MkPs are heterogeneous in MF with aberrant metabolic and inflammatory signatures
-
•
Targeting aberrant surface G6B expression may selectively ablate the MF clone
Abstract
Myelofibrosis (MF) is characterized by increased numbers of morphologically abnormal megakaryocytes (Mks). Single-cell RNA sequencing of >120,000 hematopoietic stem and progenitor cells demonstrated Mk-biased hematopoiesis across clinical and molecular MF subgroups. Mk progenitors were heterogeneous, with distinct expression of inflammatory mediators. Aberrant surface G6B expression on MF stem and progenitors could allow selective immunotherapeutic targeting of the MF clone.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Accounting for technical noise in single-cell RNA-seq experiments.
- Record: found
- Abstract: found
- Article: not found
Identification of clonogenic common lymphoid progenitors in mouse bone marrow.
- Record: found
- Abstract: found
- Article: not found
