- Record: found
- Abstract: found
- Article: found
Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor
Read this article at
Abstract
Background
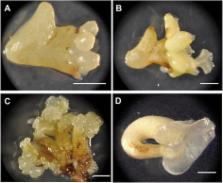
Theobroma cacao, the chocolate tree, is an important economic crop in East Africa, South East Asia, and South and Central America. Propagation of elite varieties has been achieved through somatic embryogenesis (SE) but low efficiencies and genotype dependence still presents a significant limitation for its propagation at commercial scales. Manipulation of transcription factors has been used to enhance the formation of SEs in several other plant species. This work describes the use of the transcription factor Baby Boom ( BBM) to promote the transition of somatic cacao cells from the vegetative to embryonic state.
Results
An ortholog of the Arabidopsis thaliana BBM gene ( AtBBM) was characterized in T. cacao ( TcBBM). TcBBM expression was observed throughout embryo development and was expressed at higher levels during SE as compared to zygotic embryogenesis (ZE). TcBBM overexpression in A. thaliana and T. cacao led to phenotypes associated with SE that did not require exogenous hormones. While transient ectopic expression of TcBBM provided only moderate enhancements in embryogenic potential, constitutive overexpression dramatically increased SE proliferation but also appeared to inhibit subsequent development.
Conclusion
Our work provides validation that TcBBM is an ortholog to AtBBM and has a specific role in both somatic and zygotic embryogenesis. Furthermore, our studies revealed that TcBBM transcript levels could serve as a biomarker for embryogenesis in cacao tissue. Results from transient expression of TcBBM provide confirmation that transcription factors can be used to enhance SE without compromising plant development and avoiding GMO plant production. This strategy could compliment a hormone-based method of reprogramming somatic cells and lead to more precise manipulation of SE at the regulatory level of transcription factors. The technology would benefit the propagation of elite varieties with low regeneration potential as well as the production of transgenic plants, which similarly requires somatic cell reprogramming.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
The genome of Theobroma cacao.
- Record: found
- Abstract: found
- Article: not found
A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos.
- Record: found
- Abstract: found
- Article: not found