- Record: found
- Abstract: found
- Article: found
Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species
Read this article at
Abstract
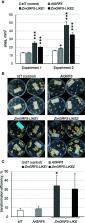
Successful regeneration of genetically modified plants from cell culture is highly dependent on the species, genotype, and tissue-type being targeted for transformation. Studies in some plant species have shown that when expression is altered, some genes regulating developmental processes are capable of triggering plant regeneration in a variety of plant cells and tissue-types previously identified as being recalcitrant to regeneration. In the present research, we report that developmental genes encoding GROWTH-REGULATING FACTORS positively enhance regeneration and transformation in both monocot and dicot species. In sugar beet ( Beta vulgaris ssp. vulgaris), ectopic expression of Arabidopsis GRF5 ( AtGRF5) in callus cells accelerates shoot formation and dramatically increases transformation efficiency. More importantly, overexpression of AtGRF5 enables the production of stable transformants in recalcitrant sugar beet varieties. The introduction of AtGRF5 and GRF5 orthologs into canola ( Brassica napus L.), soybean ( Glycine max L.), and sunflower ( Helianthus annuus L.) results in significant increases in genetic transformation of the explant tissue. A positive effect on proliferation of transgenic callus cells in canola was observed upon overexpression of GRF5 genes and AtGRF6 and AtGRF9. In soybean and sunflower, the overexpression of GRF5 genes seems to increase the proliferation of transformed cells, promoting transgenic shoot formation. In addition, the transformation of two putative AtGRF5 orthologs in maize ( Zea mays L.) significantly boosts transformation efficiency and resulted in fully fertile transgenic plants. Overall, the results suggest that overexpression of GRF genes render cells and tissues more competent to regeneration across a wide variety of crop species and regeneration processes. This sets GRFs apart from other developmental regulators and, therefore, they can potentially be applied to improve transformation of monocot and dicot plant species.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: not found
- Article: not found
Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation
- Record: found
- Abstract: found
- Article: not found