- Record: found
- Abstract: found
- Article: not found
Molecular Basis of SARS-CoV-2 Infection and Rational Design of Potential Antiviral Agents: Modeling and Simulation Approaches
Read this article at
Abstract

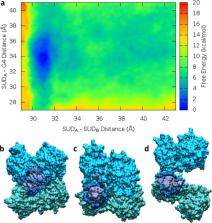
The emergence in late 2019 of the coronavirus SARS-CoV-2 has resulted in the breakthrough of the COVID-19 pandemic that is presently affecting a growing number of countries. The development of the pandemic has also prompted an unprecedented effort of the scientific community to understand the molecular bases of the virus infection and to propose rational drug design strategies able to alleviate the serious COVID-19 morbidity. In this context, a strong synergy between the structural biophysics and molecular modeling and simulation communities has emerged, resolving at the atomistic level the crucial protein apparatus of the virus and revealing the dynamic aspects of key viral processes. In this Review, we focus on how in silico studies have contributed to the understanding of the SARS-CoV-2 infection mechanism and the proposal of novel and original agents to inhibit the viral key functioning. This Review deals with the SARS-CoV-2 spike protein, including the mode of action that this structural protein uses to entry human cells, as well as with nonstructural viral proteins, focusing the attention on the most studied proteases and also proposing alternative mechanisms involving some of its domains, such as the SARS unique domain. We demonstrate that molecular modeling and simulation represent an effective approach to gather information on key biological processes and thus guide rational molecular design strategies.
Related collections
Most cited references258
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019

- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
- Record: found
- Abstract: found
- Article: not found
