- Record: found
- Abstract: found
- Article: not found
Allosteric Cross-Talk among Spike’s Receptor-Binding Domain Mutations of the SARS-CoV-2 South African Variant Triggers an Effective Hijacking of Human Cell Receptor
Read this article at
Abstract

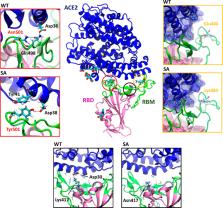
The rapid and relentless emergence of novel highly transmissible SARS-CoV-2 variants, possibly decreasing vaccine efficacy, currently represents a formidable medical and societal challenge. These variants frequently hold mutations on the Spike protein’s receptor-binding domain (RBD), which, binding to the angiotensin-converting enzyme 2 (ACE2) receptor, mediates viral entry into host cells. Here, all-atom molecular dynamics simulations and dynamical network theory of the wild-type and mutant RBD/ACE2 adducts disclose that while the N501Y mutation (UK variant) enhances the Spike’s binding affinity toward ACE2, the concomitant N501Y, E484K, and K417N mutations (South African variant) aptly adapt to increase SARS-CoV-2 propagation via a two-pronged strategy: (i) effectively grasping ACE2 through an allosteric signaling between pivotal RBD structural elements and (ii) impairing the binding of antibodies elicited by infected or vaccinated patients. This information unlocks the molecular terms and evolutionary strategies underlying the increased virulence of emerging SARS-CoV-2 variants, setting the basis for developing the next-generation anti-COVID-19 therapeutics.
Related collections
Most cited references45

- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
- Record: found
- Abstract: found
- Article: not found
Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus
- Record: found
- Abstract: found
- Article: not found
