- Record: found
- Abstract: found
- Article: found
Molecular consequences of SARS-CoV-2 liver tropism
Read this article at
Abstract
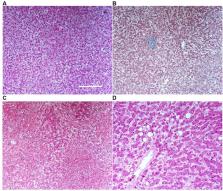
Extrapulmonary manifestations of COVID-19 have gained attention due to their links to clinical outcomes and their potential long-term sequelae 1 . Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) displays tropism towards several organs, including the heart and kidney. Whether it also directly affects the liver has been debated 2, 3 . Here we provide clinical, histopathological, molecular and bioinformatic evidence for the hepatic tropism of SARS-CoV-2. We find that liver injury, indicated by a high frequency of abnormal liver function tests, is a common clinical feature of COVID-19 in two independent cohorts of patients with COVID-19 requiring hospitalization. Using autopsy samples obtained from a third patient cohort, we provide multiple levels of evidence for SARS-CoV-2 liver tropism, including viral RNA detection in 69% of autopsy liver specimens, and successful isolation of infectious SARS-CoV-2 from liver tissue postmortem. Furthermore, we identify transcription-, proteomic- and transcription factor-based activity profiles in hepatic autopsy samples, revealing similarities to the signatures associated with multiple other viral infections of the human liver. Together, we provide a comprehensive multimodal analysis of SARS-CoV-2 liver tropism, which increases our understanding of the molecular consequences of severe COVID-19 and could be useful for the identification of organ-specific pharmacological targets.
Abstract
Wanner et al. demonstrate SARS-CoV-2 liver tropism and identify transcriptional and proteomic profiles of SARS-CoV-2 liver samples that show similarities to previously characterized hepatotropic viruses.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found