- Record: found
- Abstract: found
- Article: found
Testing, tracing and isolation in compartmental models
Abstract
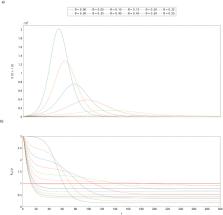
Existing compartmental mathematical modelling methods for epidemics, such as SEIR models, cannot accurately represent effects of contact tracing. This makes them inappropriate for evaluating testing and contact tracing strategies to contain an outbreak. An alternative used in practice is the application of agent- or individual-based models (ABM). However ABMs are complex, less well-understood and much more computationally expensive. This paper presents a new method for accurately including the effects of Testing, contact-Tracing and Isolation (TTI) strategies in standard compartmental models. We derive our method using a careful probabilistic argument to show how contact tracing at the individual level is reflected in aggregate on the population level. We show that the resultant SEIR-TTI model accurately approximates the behaviour of a mechanistic agent-based model at far less computational cost. The computational efficiency is such that it can be easily and cheaply used for exploratory modelling to quantify the required levels of testing and tracing, alone and with other interventions, to assist adaptive planning for managing disease outbreaks.
Author summary
The importance of modeling to inform and support decision making is widely acknowledged. Understanding how to enhance contact tracing as part of the Testing-Tracing-Isolation (TTI) strategy for mitigation of COVID is a key public policy questions. Our work develops the SEIR-TTI model as an extension of the classic Susceptible, Exposed, Infected and Recovered (SEIR) model to include tracing of contacts of people exposed to and infectious with COVID-19. We use probabilistic argument to derive contact tracing rates within a compartmental model as aggregates of contact tracing at an individual level. Our adaptation is applicable across compartmental models for infectious diseases spread. We show that our novel SEIR-TTI model can accurately approximate the behaviour of mechanistic agent-based models at far less computational cost. The SEIR-TTI model represents an important addition to the theoretical methodology of modelling infectious disease spread and we anticipate that it will be immediately applicable to the management of the COVID-19 pandemic.
Related collections
Most cited references88
- Record: found
- Abstract: found
- Article: not found
Temporal dynamics in viral shedding and transmissibility of COVID-19
- Record: found
- Abstract: not found
- Article: not found
A Contribution to the Mathematical Theory of Epidemics

- Record: found
- Abstract: found
- Article: found