- Record: found
- Abstract: found
- Article: not found
Compartmental Models of the COVID-19 Pandemic for Physicians and Physician-Scientists
review-article
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
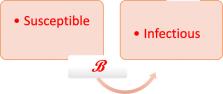
As the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection spreads globally, physicians and physician-scientists are confronted with an enlarging body of literature about the nature of this pandemic. Understanding the current epidemiological models for disease spread, mortality, and recovery is more important than ever before. One of the most relevant mathematical models relating to the spread of a pandemic is the susceptible-infectious-removed (SIR) model. Other models worth exploring are the susceptible-exposed-infectious-removed (SEIR) and the susceptible-unquarantined-quarantined-confirmed (SUQC) model.
Related collections
Most cited references3
- Record: found
- Abstract: found
- Article: not found
Modeling the epidemic dynamics and control of COVID-19 outbreak in China
Shilei Zhao, Hua. Chen (2020)

- Record: found
- Abstract: found
- Article: found
An SEIR model of influenza A virus infection and reinfection within a farrow-to-finish swine farm
- Record: found
- Abstract: found
- Article: not found
A Path-Specific SEIR Model for use with General Latent and Infectious Time Distributions
Aaron T. Porter, Jacob Oleson (2013)
