- Record: found
- Abstract: found
- Article: found
Transcriptome and Metabolome Analyses Provide Insights Into the Composition and Biosynthesis of Grassy Aroma Volatiles in White-Fleshed Pitaya

Read this article at
Abstract
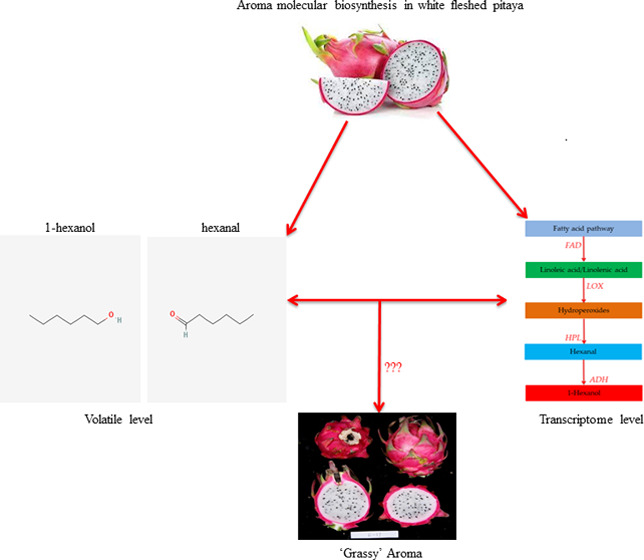
Aroma is one of the major inherent quality characteristics in fruits. Understanding the composition of aroma volatiles and their biosynthesis mechanism is crucial to improving fruit quality. However, the biosynthesis mechanism of aroma volatiles has not been characterized yet in white-fleshed pitaya ( Hylocereus undatus). This study was performed to investigate aroma volatiles and related gene expression patterns in the pulp of “mild grassy” and “strong grassy” aroma cultivars. Analysis of volatile composition and concentration showed that aldehydes, alcohols, esters, and alkenes were predominant in both cultivars. However, comparative analysis revealed a significant difference in the concentration of several metabolites, particularly hexanal and 1-hexanol. The results of the comparative transcriptome identified a large number of aroma-related differentially expressed genes. The majority of these genes were enriched in fatty acid and isoleucine degradation pathways. According to integrative analyses, changes in the expression of lipoxygenase pathway genes, specifically FAD, LOXs, HPLs, and ADHs, probably lead to the difference in strength of “grassy” aroma between both cultivars. The qRT-PCR of 18 aroma-related genes was performed to validate the transcriptome analysis. Our results identified key genes and pathways connected with the biosynthesis of aroma volatiles in white-fleshed pitaya. These results will be useful to dissect the genetic mechanism of fruit aroma in white-fleshed pitaya.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data
- Record: found
- Abstract: found
- Article: found
KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases

- Record: found
- Abstract: found
- Article: found