- Record: found
- Abstract: found
- Article: found
Metabolomics and transcriptomics analyses for characterizing the alkaloid metabolism of Chinese jujube and sour jujube fruits
Read this article at
Abstract
Introduction
Jujube is an important economic forest tree whose fruit is rich in alkaloids. Chinese jujube ( Ziziphus jujuba Mill.) and sour jujube ( Ziziphus spinosa Hu.) are the two most important species of the jujube genus. However, the mechanisms underlying the synthesis and metabolism of alkaloids in jujube fruits remain poorly understood.
Methods
In this study, the fruits of Ziziphus jujuba ‘Hupingzao’ and Ziziphus spinosa ‘Taigusuanzao’ in different harvest stages were used as test materials, we first integrated widely targeted metabolomics and transcriptomics analyses to elucidate the metabolism of alkaloids of jujube fruits.
Results
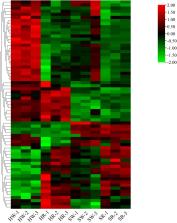
In the metabolomics analysis, 44 alkaloid metabolites were identified in 4 samples, 3 of which were unique to sour jujube fruit. The differential alkaloid metabolites (DAMs) were more accumulated in sour jujube than in Chinese jujube; further, they were more accumulated in the white ripening stage than in the red stage. DAMs were annotated to 12 metabolic pathways. Additionally, transcriptomics data revealed 259 differentially expressed genes (DEGs) involved in alkaloid synthesis and metabolism. By mapping the regulatory networks of DAMs and DEGs, we screened out important metabolites and 11 candidate genes.
Discussion
This study preliminarily elucidated the molecular mechanism of jujube alkaloid synthesis. The candidate genes regulated the synthesis of key alkaloid metabolites, but the specific regulation mechanism is unclear. Taken together, our results provide insights into the metabolic networks of alkaloid synthesis in Chinese jujube and sour jujube fruits at different harvest stages, thereby providing a theoretical reference for further research on the regulatory mechanism of jujube alkaloids and their development and utilization.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
Optimization of de novo transcriptome assembly from next-generation sequencing data.
- Record: found
- Abstract: not found
- Article: not found
A comparative metabolomics study of flavonoids in sweet potato with different flesh colors ( Ipomoea batatas (L.) Lam)
- Record: found
- Abstract: found
- Article: not found