- Record: found
- Abstract: found
- Article: found
Roles and Programming of Arabidopsis ARGONAUTE Proteins during Turnip Mosaic Virus Infection
Read this article at
Abstract
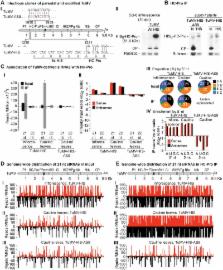
In eukaryotes, ARGONAUTE proteins (AGOs) associate with microRNAs (miRNAs), short interfering RNAs (siRNAs), and other classes of small RNAs to regulate target RNA or target loci. Viral infection in plants induces a potent and highly specific antiviral RNA silencing response characterized by the formation of virus-derived siRNAs. Arabidopsis thaliana has ten AGO genes of which AGO1, AGO2, and AGO7 have been shown to play roles in antiviral defense. A genetic analysis was used to identify and characterize the roles of AGO proteins in antiviral defense against Turnip mosaic virus (TuMV) in Arabidopsis. AGO1, AGO2 and AGO10 promoted anti-TuMV defense in a modular way in various organs, with AGO2 providing a prominent antiviral role in leaves. AGO5, AGO7 and AGO10 had minor effects in leaves. AGO1 and AGO10 had overlapping antiviral functions in inflorescence tissues after systemic movement of the virus, although the roles of AGO1 and AGO10 accounted for only a minor amount of the overall antiviral activity. By combining AGO protein immunoprecipitation with high-throughput sequencing of associated small RNAs, AGO2, AGO10, and to a lesser extent AGO1 were shown to associate with siRNAs derived from silencing suppressor (HC-Pro)-deficient TuMV-AS9, but not with siRNAs derived from wild-type TuMV. Co-immunoprecipitation and small RNA sequencing revealed that viral siRNAs broadly associated with wild-type HC-Pro during TuMV infection. These results support the hypothesis that suppression of antiviral silencing during TuMV infection, at least in part, occurs through sequestration of virus-derived siRNAs away from antiviral AGO proteins by HC-Pro. These findings indicate that distinct AGO proteins function as antiviral modules, and provide a molecular explanation for the silencing suppressor activity of HC-Pro.
Author Summary
RNA silencing is a primary, adaptive defense system against viruses in plants. Viruses have evolved counter-defensive mechanisms that inhibit RNA silencing through the activity of silencing suppressor proteins. Understanding how antiviral silencing is controlled, and how suppressor proteins function, is essential for understanding how plants normally resist viruses, why some viruses are highly virulent in different hosts, and how sustainable antiviral resistance strategies can be deployed in agricultural settings. We used a mutant version of Turnip mosaic virus lacking a functional silencing suppressor (HC-Pro) to understand the genetic requirements for resistance in the model plant Arabidopsis thaliana. We focused on ARGONAUTE proteins, which have long been hypothesized to bind short interfering RNAs (siRNAs) derived from virus genomes for use as sequence-specific guides to recognize and target viral RNA for degradation or repression. We demonstrated specialized antiviral roles for specific ARGONAUTES and showed that several can bind viral siRNAs from across the entire viral genome. However, ARGONAUTE proteins are only loaded with virus-derived siRNAs in the absence of HC-Pro, which we showed binds siRNAs from the viral genome. This indicates that several AGO proteins, which collectively are necessary for full anti-TuMV defense, need to properly load virus-derived siRNAs to execute their antiviral roles.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Genome-wide insertional mutagenesis of Arabidopsis thaliana.
- Record: found
- Abstract: found
- Article: not found
Widespread translational inhibition by plant miRNAs and siRNAs.
- Record: found
- Abstract: found
- Article: not found