- Record: found
- Abstract: found
- Article: found
Presence of Mobile Tigecycline Resistance Gene tet(X4) in Clinical Klebsiella pneumoniae
Read this article at
ABSTRACT
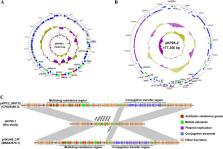
The recently emerged plasmid-mediated tigecycline resistance gene tet(X4) has mainly been detected in Escherichia coli but never in Klebsiella pneumoniae. Herein, we identified a clinical K. pneumoniae isolate that harbored the tet(X4) gene located on a non-self-transferable IncFII-type plasmid, which could be cotransferred with a conjugative plasmid to E. coli C600. The extending of bacterial species carrying tet(X4) suggested the increasing risk of spreading mobile tigecycline resistance genes among important pathogens in clinical settings.
IMPORTANCE Tigecycline, the first member of glycylcycline class antibiotic, is often considered one of the effective antibiotics against multidrug-resistant (MDR) infections. However, the emergence and wide distribution of two novel plasmid-mediated tigecycline resistance genes, tet(X3) and tet(X4), pose a great threat to the clinical use of tigecycline. The newly tet(X) variants have been identified from multiple different bacterial species, but the tet(X) variant in the Klebsiella pneumoniae strain has been reported only once before. In this study, we identified a clinical K. pneumoniae isolate that harbored a non-self-transferable tet(X4)-carrying plasmid. This plasmid has never been found in other tet(X4)-harboring strains and could be cotransferred with a conjugative plasmid to the recipient strain. Our findings indicate that the tet(X4) gene breaks through its original bacterial species and spreads to some important nosocomial pathogens, which posed a serious threat to public health.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans
- Record: found
- Abstract: found
- Article: not found