- Record: found
- Abstract: found
- Article: found
Comprehensive Genomic Investigation of Tigecycline Resistance Gene tet(X4)-Bearing Strains Expanding among Different Settings
Read this article at
ABSTRACT
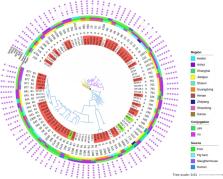
The emergence of plasmid-mediated tigecycline resistance genes has attracted a great deal of attention globally. Currently, no comprehensive in-depth genomic epidemiology study of tet(X4)-bearing pathogens present of pork origin as the One Health approach has been performed. Herein, 139 fresh pork samples were collected from multiple regions in China and 58 tet(X4)-positive strains were identified. The tet(X4) gene mainly distributed in Escherichia coli ( n = 55). Besides, 4 novel tet(X4)-positive bacterial species Klebsiella pneumoniae ( n = 2), Klebsiella quasipneumoniae ( n = 1), Citrobacter braakii ( n = 1) and Citrobacter freundii ( n = 1) were first characterized here. Four different core tet(X4)-bearing genetic environments and five types of tet(X4)-bearing tandem duplications were discovered among 58 strains. The results of the phylogenetic tree showed that there was some correlation between E. coli strains from pork, human, pig farms, and slaughterhouses. A total of seven types of plasmid replicons were found in tet(X4)-positive plasmids, among which multireplicon plasmids were observed. Notably, two tet(X4)-positive fusion plasmids pCSZ11R (IncX1-IncFIA-IncFIB-IncFIC) and pCSX5G-tetX4 (IncX1-IncFII-IncFIA) were formed by IS 26 in the hot spot. Besides, six samples were identified to harbor two different tet(X4)-bearing strains. More interestingly, the absolute quantitative results showed that the expression levels of tet(X4) between different strains with different tet(X4) copies were approximate. In this study, the genetic environment of tet(X4)-positive plasmids containing different plasmid replicons was analyzed to provide a basis for the further development of effective control measures. It is also highlighted that animal-borne tet(X4)-bearing pathogens incur a transmission risk to consumed food. Therefore, there is an urgent need for large-scale monitoring as well as the development of effective control measures.
IMPORTANCE Tigecycline was considered the last-line drug against serious infections caused by multidrug-resistant Gram-negative bacteria. However, the plasmid-mediated tigecycline resistance gene tet(X) has been widely reported in different sources of Enterobacterales and Acinetobacter in China. China is one of the largest pig-producing nations in the world, and in-depth investigation of gene in pork is vital to figure out the fundamental dissemination of these genes and set up a reasonable control framework. In this study, we conducted an in-depth and systematic analysis of the diversity of tet(X4)-positive plasmids and the genetic environment of tet(X4) contained in pork samples from multiple regions of China, providing a basis for further development of effective control measures. It is also highlighted that animal-borne tet(X4)-bearing pathogens incur a transmission risk to consumed food. Therefore, there is an urgent need for large-scale monitoring as well as the development of effective control measures.
Related collections
Most cited references41

- Record: found
- Abstract: found
- Article: found
Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads
- Record: found
- Abstract: found
- Article: not found
Assembly of long, error-prone reads using repeat graphs
- Record: found
- Abstract: found
- Article: found