- Record: found
- Abstract: found
- Article: found
Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study
Read this article at
Abstract
Background
Non-small-cell lung cancer (NSCLC) patients often demonstrate varying clinical courses and outcomes, even within the same tumor stage. This study explores deep learning applications in medical imaging allowing for the automated quantification of radiographic characteristics and potentially improving patient stratification.
Methods and findings
We performed an integrative analysis on 7 independent datasets across 5 institutions totaling 1,194 NSCLC patients (age median = 68.3 years [range 32.5–93.3], survival median = 1.7 years [range 0.0–11.7]). Using external validation in computed tomography (CT) data, we identified prognostic signatures using a 3D convolutional neural network (CNN) for patients treated with radiotherapy ( n = 771, age median = 68.0 years [range 32.5–93.3], survival median = 1.3 years [range 0.0–11.7]). We then employed a transfer learning approach to achieve the same for surgery patients ( n = 391, age median = 69.1 years [range 37.2–88.0], survival median = 3.1 years [range 0.0–8.8]). We found that the CNN predictions were significantly associated with 2-year overall survival from the start of respective treatment for radiotherapy (area under the receiver operating characteristic curve [AUC] = 0.70 [95% CI 0.63–0.78], p < 0.001) and surgery (AUC = 0.71 [95% CI 0.60–0.82], p < 0.001) patients. The CNN was also able to significantly stratify patients into low and high mortality risk groups in both the radiotherapy ( p < 0.001) and surgery ( p = 0.03) datasets. Additionally, the CNN was found to significantly outperform random forest models built on clinical parameters—including age, sex, and tumor node metastasis stage—as well as demonstrate high robustness against test–retest (intraclass correlation coefficient = 0.91) and inter-reader (Spearman’s rank-order correlation = 0.88) variations. To gain a better understanding of the characteristics captured by the CNN, we identified regions with the most contribution towards predictions and highlighted the importance of tumor-surrounding tissue in patient stratification. We also present preliminary findings on the biological basis of the captured phenotypes as being linked to cell cycle and transcriptional processes. Limitations include the retrospective nature of this study as well as the opaque black box nature of deep learning networks.
Conclusions
Our results provide evidence that deep learning networks may be used for mortality risk stratification based on standard-of-care CT images from NSCLC patients. This evidence motivates future research into better deciphering the clinical and biological basis of deep learning networks as well as validation in prospective data.
Abstract
Hugo Aerts and colleagues evaluate the ability of deep learning networks to extract relevant features from computed tomography lung cancer images and stratify patients into low and high mortality risk groups.
Author summary
Why was this study done?
-
Cancer is one of the leading causes of death worldwide, with lung cancer being the second most commonly diagnosed cancer in both men and women in the US.
-
Prognosis in lung cancer patients is primarily determined through tumor staging, which in turn is based on a relatively coarse and discrete stratification.
-
Radiographic medical images offer patient- and tumor-specific information that could be used to complement clinical prognostic evaluation efforts.
-
Recent advances in radiomics through applications of artificial intelligence, computer vision, and deep learning allow for the extraction and mining of numerous quantitative features from radiographic images.
What did the researchers do and find?
-
We designed an analysis setup comprising 7 independent datasets across 5 institutions totaling 1,194 patients with non-small-cell lung cancer imaged with computed tomography and treated with either radiotherapy or surgery.
-
We evaluated the prognostic signatures of quantitative imaging features extracted through deep learning networks, and assessed their ability to stratify patients into low and high mortality risk groups as per a 2-year overall survival cutoff.
-
In patients treated with surgery, deep learning networks significantly outperformed models based on predefined tumor features as well as tumor volume and maximum diameter.
-
In addition to highlighting image regions with prognostic influence, we evaluated the deep learning features for robustness against physiological imaging artifacts and input variability, as well as correlated them with molecular information through gene expression data.
What do these findings mean?
-
We found that deep learning features significantly outperform existing prognostication methods in surgery patients, hinting at their utility in patient stratification and potentially sparing low mortality risk groups from adjuvant chemotherapy.
-
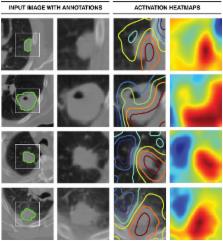
We demonstrated that areas within and beyond the tumor—especially the tumor–stroma interfaces—had the largest contributions to the prognostic signature, highlighting the importance of tumor-surrounding tissue in patient stratification.
-
Preliminary genomic associations in this study suggest correlations between the deep learning feature representations and cell cycle and transcriptional processes.
-
Despite their obscure inner workings and lack of a strong theoretical backing, deep learning networks demonstrate a prognostic signal and robustness against specific noise artifacts. This finding motivates further prospective studies validating their utility in patient stratification and the development of personalized cancer treatment plans.
Related collections
Most cited references39
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing

- Record: found
- Abstract: found
- Article: found
Machine Learning methods for Quantitative Radiomic Biomarkers

- Record: found
- Abstract: found
- Article: found