- Record: found
- Abstract: found
- Article: found
Unlocking the potential of T‐cell metabolism reprogramming: Advancing single‐cell approaches for precision immunotherapy in tumour immunity
Read this article at
Abstract
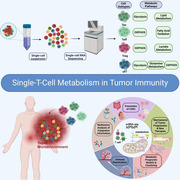
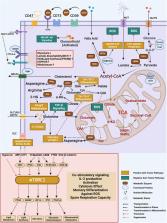
As single‐cell RNA sequencing enables the detailed clustering of T‐cell subpopulations and facilitates the analysis of T‐cell metabolic states and metabolite dynamics, it has gained prominence as the preferred tool for understanding heterogeneous cellular metabolism. Furthermore, the synergistic or inhibitory effects of various metabolic pathways within T cells in the tumour microenvironment are coordinated, and increased activity of specific metabolic pathways generally corresponds to increased functional activity, leading to diverse T‐cell behaviours related to the effects of tumour immune cells, which shows the potential of tumour‐specific T cells to induce persistent immune responses. A holistic understanding of how metabolic heterogeneity governs the immune function of specific T‐cell subsets is key to obtaining field‐level insights into immunometabolism. Therefore, exploring the mechanisms underlying the interplay between T‐cell metabolism and immune functions will pave the way for precise immunotherapy approaches in the future, which will empower us to explore new methods for combating tumours with enhanced efficacy.
Abstract
Single T‐cell metabolism in tumour immunity
-
Single‐cell RNA sequencing has gained prominence as the preferred tool for clarifying cell subtypes and metabolic heterogeneity.
-
Various metabolic pathways within T cells in the tumour microenvironment are coordinated, leading to diverse T‐cell behaviours.
-
Understanding the mechanisms underlying the interplay between T‐cell metabolism and immune functions will pave the way for precise immunotherapy approaches.
Related collections
Most cited references238

- Record: found
- Abstract: found
- Article: found
Integrated analysis of multimodal single-cell data
- Record: found
- Abstract: found
- Article: not found
Elements of cancer immunity and the cancer–immune set point
- Record: found
- Abstract: found
- Article: not found