- Record: found
- Abstract: found
- Article: found
SARS-CoV-2 requires acidic pH to infect cells
Read this article at
Significance
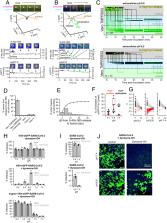
Infection by SARS-CoV-2 depends upon the large spike (S) protein decorating the virions and is responsible for receptor engagement and subsequent fusion of viral and cellular membranes allowing release of virion contents into the cell. Using new single-particle imaging tools to visualize and track the successive steps from virion attachment to fusion, combined with chemical and genetic perturbations of the cells, we provide direct evidence for the cellular uptake routes of productive infection in multiple cell types and their dependence on proteolysis of S by cell surface or endosomal proteases. We show that fusion and content release always require the acidic environment from endosomes, preceded by liberation of the S1 fragment which depends on angiotensin converting enzyme receptor engagement.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cell entry starts with membrane attachment and ends with spike (S) protein–catalyzed membrane fusion depending on two cleavage steps, namely, one usually by furin in producing cells and the second by TMPRSS2 on target cells. Endosomal cathepsins can carry out both. Using real-time three-dimensional single-virion tracking, we show that fusion and genome penetration require virion exposure to an acidic milieu of pH 6.2 to 6.8, even when furin and TMPRSS2 cleavages have occurred. We detect the sequential steps of S1-fragment dissociation, fusion, and content release from the cell surface in TMPRRS2-overexpressing cells only when exposed to acidic pH. We define a key role of an acidic environment for successful infection, found in endosomal compartments and at the surface of TMPRSS2-expressing cells in the acidic milieu of the nasal cavity.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor

- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation

- Record: found
- Abstract: found
- Article: found