- Record: found
- Abstract: found
- Article: found
dRFEtools: dynamic recursive feature elimination for omics
Read this article at
Abstract
Motivation
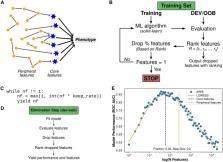
Advances in technology have generated larger omics datasets with potential applications for machine learning. In many datasets, however, cost and limited sample availability result in an excessively higher number of features as compared to observations. Moreover, biological processes are associated with networks of core and peripheral genes, while traditional feature selection approaches capture only core genes.
Results
To overcome these limitations, we present dRFEtools that implements dynamic recursive feature elimination (RFE), reducing computational time with high accuracy compared to standard RFE, expanding dynamic RFE to regression algorithms, and outputting the subsets of features that hold predictive power with and without peripheral features. dRFEtools integrates with scikit-learn (the popular Python machine learning platform) and thus provides new opportunities for dynamic RFE in large-scale omics data while enhancing its interpretability.
Availability and implementation
dRFEtools is freely available on PyPI at https://pypi.org/project/drfetools/ or on GitHub https://github.com/LieberInstitute/dRFEtools, implemented in Python 3, and supported on Linux, Windows, and Mac OS.
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
An Expanded View of Complex Traits: From Polygenic to Omnigenic
- Record: found
- Abstract: found
- Article: not found
Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis
- Record: found
- Abstract: found
- Article: not found