- Record: found
- Abstract: found
- Article: found
Sex affects transcriptional associations with schizophrenia across the dorsolateral prefrontal cortex, hippocampus, and caudate nucleus
Read this article at
Abstract
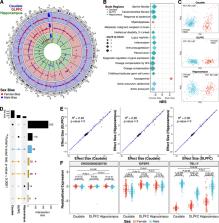
Schizophrenia is a complex neuropsychiatric disorder with sexually dimorphic features, including differential symptomatology, drug responsiveness, and male incidence rate. Prior large-scale transcriptome analyses for sex differences in schizophrenia have focused on the prefrontal cortex. Analyzing BrainSeq Consortium data (caudate nucleus: n = 399, dorsolateral prefrontal cortex: n = 377, and hippocampus: n = 394), we identified 831 unique genes that exhibit sex differences across brain regions, enriched for immune-related pathways. We observed X-chromosome dosage reduction in the hippocampus of male individuals with schizophrenia. Our sex interaction model revealed 148 junctions dysregulated in a sex-specific manner in schizophrenia. Sex-specific schizophrenia analysis identified dozens of differentially expressed genes, notably enriched in immune-related pathways. Finally, our sex-interacting expression quantitative trait loci analysis revealed 704 unique genes, nine associated with schizophrenia risk. These findings emphasize the importance of sex-informed analysis of sexually dimorphic traits, inform personalized therapeutic strategies in schizophrenia, and highlight the need for increased female samples for schizophrenia analyses.
Abstract
Schizophrenia research has traditionally overlooked sex differences. Here, the authors show the importance of sex-based analysis across multi-brain regions by identifying sex-specific genes and genetic interactions in schizophrenia and sex-specific risk.
Related collections
Most cited references96
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found