- Record: found
- Abstract: found
- Article: found
Kaempferol Alleviates Murine Experimental Colitis by Restoring Gut Microbiota and Inhibiting the LPS-TLR4-NF-κB Axis
Read this article at
Abstract
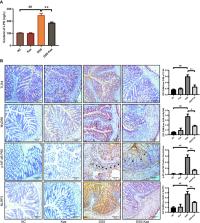
Intestinal microbiota dysbiosis is an established characteristic of ulcerative colitis (UC). Regulating the gut microbiota is an attractive alternative UC treatment strategy, considering the potential adverse effects of synthetic drugs used to treat UC. Kaempferol (Kae) is an anti-inflammatory and antioxidant flavonoid derived from a variety of medicinal plants. In this study, we determined the efficacy and mechanism of action of Kae as an anti-UC agent in dextran sulfate sodium (DSS)-induced colitis mice. DSS challenge in a mouse model of UC led to weight loss, diarrhea accompanied by mucous and blood, histological abnormalities, and shortening of the colon, all of which were significantly alleviated by pretreatment with Kae. In addition, intestinal permeability was shown to improve using fluorescein isothiocyanate (FITC)–dextran administration. DSS-induced destruction of the intestinal barrier was also significantly prevented by Kae administration via increases in the levels of ZO-1, occludin, and claudin-1. Furthermore, Kae pretreatment decreased the levels of IL-1β, IL-6, and TNF-α and downregulated transcription of an array of inflammatory signaling molecules, while it increased IL-10 mRNA expression. Notably, Kae reshaped the intestinal microbiome by elevating the Firmicutes to Bacteroidetes ratio; increasing the linear discriminant analysis scores of beneficial bacteria, such as Prevotellaceae and Ruminococcaceae; and reducing the richness of Proteobacteria in DSS-challenged mice. There was also an evident shift in the profile of fecal metabolites in the Kae treatment group. Serum LPS levels and downstream TLR4-NF-κB signaling were downregulated by Kae supplementation. Moreover, fecal microbiota transplantation from Kae-treated mice to the DSS-induced mice confirmed the effects of Kae on modulating the gut microbiota to alleviate UC. Therefore, Kae may exert protective effects against colitis mice through regulating the gut microbiota and TLR4-related signaling pathways. This study demonstrates the anti-UC effects of Kae and its potential therapeutic mechanisms, and offers novel insights into the prevention of inflammatory diseases using natural products.
Related collections
Most cited references63

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.

- Record: found
- Abstract: found
- Article: found