- Record: found
- Abstract: found
- Article: found
Diet-Induced Alterations in Total and Metabolically Active Microbes within the Rumen of Dairy Cows
Read this article at
Abstract
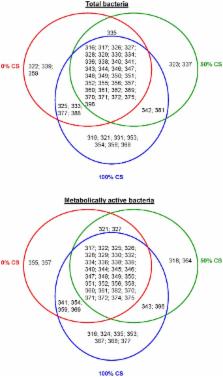
DNA-based techniques are widely used to study microbial populations; however, this approach is not specific to active microbes, because DNA may originate from inactive and/or dead cells. Using cDNA and DNA, respectively, we aimed to discriminate the active microbes from the total microbial community within the rumen of dairy cows fed diets with increasing proportions of corn silage (CS). Nine multiparous lactating Holstein cows fitted with ruminal cannulas were used in a replicated 3×3 Latin square (32-d period; 21-d adaptation) design to investigate diet-induced shifts in microbial populations by targeting the rDNA gene. Cows were fed a total mixed ration with the forage portion being either barley silage (0% CS), a 50∶50 mixture of barley silage and corn silage (50% CS), or corn silage (100% CS). No differences were found for total microbes analyzed by quantitative PCR, but changes were observed within the active ones. Feeding more CS to dairy cows was accompanied by an increase in Prevotella rRNA transcripts ( P = 0.10) and a decrease in the protozoal rRNA transcripts ( P<0.05). Although they were distributed differently among diets, 78% of the amplicons detected in DNA- and cDNA-based fingerprints were common to total and active bacterial communities. These may represent a bacterial core of abundant and active cells that drive the fermentation processes. In contrast, 10% of amplicons were specific to total bacteria and may represent inactive or dead cells, whereas 12% were only found within the active bacterial community and may constitute slow-growing bacteria with high metabolic activity. It appears that cDNA-based analysis is more discriminative to identify diet-induced shifts within the microbial community. This approach allows the detection of diet-induced changes in the microbial populations as well as particular bacterial amplicons that remained undetected using DNA-based methods.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
Phylogenetic structure of the prokaryotic domain: the primary kingdoms.
- Record: found
- Abstract: found
- Article: not found
Methane mitigation in ruminants: from microbe to the farm scale.
- Record: found
- Abstract: found
- Article: not found