- Record: found
- Abstract: found
- Article: found
Transcription-dependent targeting of Hda1C to hyperactive genes mediates H4-specific deacetylation in yeast
Read this article at
Abstract
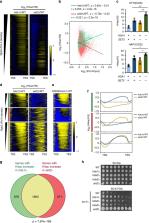
In yeast, Hda1 histone deacetylase complex (Hda1C) preferentially deacetylates histones H3 and H2B, and functionally interacts with Tup1 to repress transcription. However, previous studies identified global increases in histone H4 acetylation in cells lacking Hda1, a component of Hda1C. Here, we find that Hda1C binds to hyperactive genes, likely via the interaction between the Arb2 domain of Hda1 and RNA polymerase II. Additionally, we report that Hda1C specifically deacetylates H4, but not H3, at hyperactive genes to partially inhibit elongation. This role is contrast to that of the Set2–Rpd3S pathway deacetylating histones at infrequently transcribed genes. We also find that Hda1C deacetylates H3 at inactive genes to delay the kinetics of gene induction. Therefore, in addition to fine-tuning of transcriptional response via H3-specific deacetylation, Hda1C may modulate elongation by specifically deacetylating H4 at highly transcribed regions.
Abstract
In yeast, Hda1 histone deacetylase complex (Hda1C) preferentially deacetylates H3 and H2B, but has also been implicated in global H4 deacetylation. Here, the authors provide evidence that Hda1C binds to hyperactive genes, where it specifically deacetylates H4 to partially inhibit elongation, suggesting a role for Hda1C in elongation by specifically deacetylating H4 at highly transcribed regions.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes.
- Record: found
- Abstract: found
- Article: not found
Histone exchange, chromatin structure and the regulation of transcription.
- Record: found
- Abstract: found
- Article: not found