- Record: found
- Abstract: found
- Article: found
Molecular-scale visualization of sarcomere contraction within native cardiomyocytes
Read this article at
Abstract
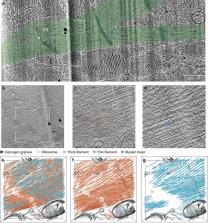
Sarcomeres, the basic contractile units of striated muscle, produce the forces driving muscular contraction through cross-bridge interactions between actin-containing thin filaments and myosin II-based thick filaments. Until now, direct visualization of the molecular architecture underlying sarcomere contractility has remained elusive. Here, we use in situ cryo-electron tomography to unveil sarcomere contraction in frozen-hydrated neonatal rat cardiomyocytes. We show that the hexagonal lattice of the thick filaments is already established at the neonatal stage, with an excess of thin filaments outside the trigonal positions. Structural assessment of actin polarity by subtomogram averaging reveals that thin filaments in the fully activated state form overlapping arrays of opposite polarity in the center of the sarcomere. Our approach provides direct evidence for thin filament sliding during muscle contraction and may serve as a basis for structural understanding of thin filament activation and actomyosin interactions inside unperturbed cellular environments.
Abstract
Sarcomeres, the building blocks of striated muscles, comprise ordered actomyosin arrays involved in force production. Here, the authors visualize sarcomere organization in neonatal cardiomyocytes with in situ cryo-electron tomography, revealing a reduced order of the thin filaments, their sliding and functional states enabling contraction.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: found
- Article: not found
MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy
- Record: found
- Abstract: found
- Article: not found