- Record: found
- Abstract: found
- Article: found
A Model for a Chikungunya Outbreak in a Rural Cambodian Setting: Implications for Disease Control in Uninfected Areas
Read this article at
Abstract
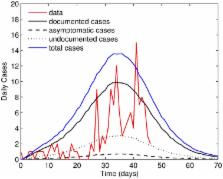
Following almost 30 years of relative silence, chikungunya fever reemerged in Kenya in 2004. It subsequently spread to the islands of the Indian Ocean, reaching Southeast Asia in 2006. The virus was first detected in Cambodia in 2011 and a large outbreak occurred in the village of Trapeang Roka Kampong Speu Province in March 2012, in which 44% of the villagers had a recent infection biologically confirmed. The epidemic curve was constructed from the number of biologically-confirmed CHIKV cases per day determined from the date of fever onset, which was self-reported during a data collection campaign conducted in the village after the outbreak. All individuals participating in the campaign had infections confirmed by laboratory analysis, allowing for the identification of asymptomatic cases and those with an unreported date of fever onset. We develop a stochastic model explicitly including such cases, all of whom do not appear on the epidemic curve. We estimate the basic reproduction number of the outbreak to be 6.46 (95% C.I. [6.24, 6.78]). We show that this estimate is particularly sensitive to changes in the biting rate and mosquito longevity. Our model also indicates that the infection was more widespread within the population on the reported epidemic start date. We show that the exclusion of asymptomatic cases and cases with undocumented onset dates can lead to an underestimation of the reproduction number which, in turn, could negatively impact control strategies implemented by public health authorities. We highlight the need for properly documenting newly emerging pathogens in immunologically naive populations and the importance of identifying the route of disease introduction.
Author Summary
During the recent resurgence of chikungunya, the scale of imported cases into previously unaffected countries has caused great concern due to the presence of a competent vector ( Aedes albopictus) in many of these regions. This study describes a mathematical model for a chikungunya outbreak in the rural Cambodian village of Trapeang Roka, where a chikungunya epidemic was recorded and documented in March 2012. The outbreak data is unique, in that all infections were confirmed by laboratory analysis, enabling the identification of asymptomatic individuals, in addition to individuals who failed to report details of their infection. A stochastic model, partitioning the infectious population into three distinct classes, is implemented using Gillespie's algorithm. We show that the incorporation of both biologically-confirmed symptomatic cases undocumented by date of fever onset and asymptomatic cases yields a higher estimate of the reproduction number. Our results highlight how reproduction numbers could be underestimated by limiting analysis to the epidemic curve. Carefully documenting cases and performing laboratory testing in cluster regions, such as the village considered here, could provide a more comprehensive insight into the true infection dynamics.
Related collections
Most cited references59
- Record: found
- Abstract: not found
- Article: not found
Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus.
- Record: found
- Abstract: found
- Article: not found
Two Chikungunya Isolates from the Outbreak of La Reunion (Indian Ocean) Exhibit Different Patterns of Infection in the Mosquito, Aedes albopictus
- Record: found
- Abstract: found
- Article: not found