- Record: found
- Abstract: found
- Article: found
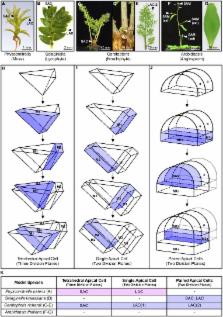
Ferns: the missing link in shoot evolution and development
Read this article at
Abstract
Shoot development in land plants is a remarkably complex process that gives rise to an extreme diversity of forms. Our current understanding of shoot developmental mechanisms comes almost entirely from studies of angiosperms (flowering plants), the most recently diverged plant lineage. Shoot development in angiosperms is based around a layered multicellular apical meristem that produces lateral organs and/or secondary meristems from populations of founder cells at its periphery. In contrast, non-seed plant shoots develop from either single apical initials or from a small population of morphologically distinct apical cells. Although developmental and molecular information is becoming available for non-flowering plants, such as the model moss Physcomitrella patens, making valid comparisons between highly divergent lineages is extremely challenging. As sister group to the seed plants, the monilophytes (ferns and relatives) represent an excellent phylogenetic midpoint of comparison for unlocking the evolution of shoot developmental mechanisms, and recent technical advances have finally made transgenic analysis possible in the emerging model fern Ceratopteris richardii. This review compares and contrasts our current understanding of shoot development in different land plant lineages with the aim of highlighting the potential role that the fern C. richardii could play in shedding light on the evolution of underlying genetic regulatory mechanisms.
Related collections
Most cited references152
- Record: found
- Abstract: found
- Article: not found
The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants.
- Record: found
- Abstract: found
- Article: not found
The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes.
- Record: found
- Abstract: found
- Article: not found