- Record: found
- Abstract: found
- Article: found
Single-cell RNA-sequencing data analysis reveals a highly correlated triphasic transcriptional response to SARS-CoV-2 infection
Read this article at
Abstract
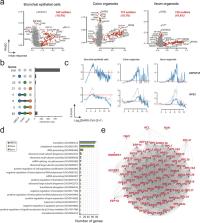
Single-cell RNA sequencing (scRNA-seq) is currently one of the most powerful techniques available to study the transcriptional response of thousands of cells to an external perturbation. Here, we perform a pseudotime analysis of SARS-CoV-2 infection using publicly available scRNA-seq data from human bronchial epithelial cells and colon and ileum organoids. Our results reveal that, for most genes, the transcriptional response to SARS-CoV-2 infection follows a non-linear pattern characterized by an initial and a final down-regulatory phase separated by an intermediate up-regulatory stage. A correlation analysis of transcriptional profiles suggests a common mechanism regulating the mRNA levels of most genes. Interestingly, genes encoded in the mitochondria or involved in translation exhibited distinct pseudotime profiles. To explain our results, we propose a simple model where nuclear export inhibition of nsp1-sensitive transcripts will be sufficient to explain the transcriptional shutdown of SARS-CoV-2 infected cells.
Abstract
A pseudotime analysis of the host cell response to SARS-CoV-2 infection using publicly available scRNA-seq data derived from in vitro studies reveals a non-lineal, triphasic transcriptional response and suggests a global regulatory mechanism.
Related collections
Most cited references77
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing

- Record: found
- Abstract: found
- Article: found