- Record: found
- Abstract: found
- Article: found
Cross-resistance is modular in bacteria–phage interactions
Read this article at
Abstract
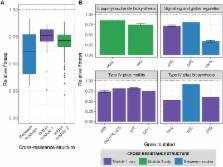
Phages shape the structure of natural bacterial communities and can be effective therapeutic agents. Bacterial resistance to phage infection, however, limits the usefulness of phage therapies and could destabilise community structures, especially if individual resistance mutations provide cross-resistance against multiple phages. We currently understand very little about the evolution of cross-resistance in bacteria–phage interactions. Here we show that the network structure of cross-resistance among spontaneous resistance mutants of Pseudomonas aeruginosa evolved against each of 27 phages is highly modular. The cross-resistance network contained both symmetric (reciprocal) and asymmetric (nonreciprocal) cross-resistance, forming two cross-resistance modules defined by high within- but low between-module cross-resistance. Mutations conferring cross-resistance within modules targeted either lipopolysaccharide or type IV pilus biosynthesis, suggesting that the modularity of cross-resistance was structured by distinct phage receptors. In contrast, between-module cross-resistance was provided by mutations affecting the alternative sigma factor, RpoN, which controls many lifestyle-associated functions, including motility, biofilm formation, and quorum sensing. Broader cross-resistance range was not associated with higher fitness costs or weaker resistance against the focal phage used to select resistance. However, mutations in rpoN, providing between-module cross-resistance, were associated with higher fitness costs than mutations associated with within-module cross-resistance, i.e., in genes encoding either lipopolysaccharide or type IV pilus biosynthesis. The observed structure of cross-resistance predicted both the frequency of resistance mutations and the ability of phage combinations to suppress bacterial growth. These findings suggest that the evolution of cross-resistance is common, is likely to play an important role in the dynamic structure of bacteria–phage communities, and could inform the design principles for phage therapy treatments.
Author summary
Phage therapy is a promising alternative to antibiotics for treating bacterial infections. Yet as with antibiotics, bacteria readily evolve resistance to phage attack, including cross-resistance that protects against multiple phages at once and so limits the usefulness of phage cocktails. Here we show, using laboratory experimental evolution of resistance against 27 phages in P. aeruginosa, that cross-resistance is common and determines the ability of phage combinations to suppress bacterial growth. Using whole-genome sequencing, we show that cross-resistance is most common against multiple phages that use the same receptor but that global regulator mutations provide generalist resistance, probably by simultaneously affecting the expression of multiple different phage receptors. Future trials should test if these features of cross-resistance evolution translate to more complex in vivo environments and can therefore be exploited to design more effective phage therapies for the clinic.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: found
Community structure in social and biological networks
- Record: found
- Abstract: found
- Article: not found
Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients.
- Record: found
- Abstract: found
- Article: not found