- Record: found
- Abstract: found
- Article: found
Fertilization Shapes Bacterial Community Structure by Alteration of Soil pH
Read this article at
Abstract
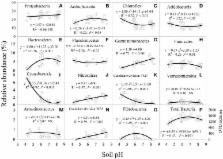
Application of chemical fertilizer or manure can affect soil microorganisms directly by supplying nutrients and indirectly by altering soil pH. However, it remains uncertain which effect mostly shapes microbial community structure. We determined soil bacterial diversity and community structure by 454 pyrosequencing the V1-V3 regions of 16S rRNA genes after 7-years (2007–2014) of applying chemical nitrogen, phosphorus and potassium (NPK) fertilizers, composted manure or their combination to acidic (pH 5.8), near-neutral (pH 6.8) or alkaline (pH 8.4) Eutric Regosol soil in a maize-vegetable rotation in southwest China. In alkaline soil, nutrient sources did not affect bacterial Operational Taxonomic Unit (OTU) richness or Shannon diversity index, despite higher available N, P, K, and soil organic carbon in fertilized than in unfertilized soil. In contrast, bacterial OTU richness and Shannon diversity index were significantly lower in acidic and near-neutral soils under NPK than under manure or their combination, which corresponded with changes in soil pH. Permutational multivariate analysis of variance showed that bacterial community structure was significantly affected across these three soils, but the PCoA ordination patterns indicated the effect was less distinct among nutrient sources in alkaline than in acidic and near-neural soils. Distance-based redundancy analysis showed that bacterial community structures were significantly altered by soil pH in acidic and near-neutral soils, but not by any soil chemical properties in alkaline soil. The relative abundance (%) of most bacterial phyla was higher in near-neutral than in acidic or alkaline soils. The most dominant phyla were Proteobacteria (24.6%), Actinobacteria (19.7%), Chloroflexi (15.3%) and Acidobacteria (12.6%); the medium dominant phyla were Bacterioidetes (5.3%), Planctomycetes (4.8%), Gemmatimonadetes (4.5%), Firmicutes (3.4%), Cyanobacteria (2.1%), Nitrospirae (1.8%), and candidate division TM7 (1.0%); the least abundant phyla were Verrucomicrobia (0.7%), Armatimonadetes (0.6%), candidate division WS3 (0.4%) and Fibrobacteres (0.3%). In addition, Cyanobacteria and candidate division TM7 were more abundant in acidic soil, whereas Gemmatimonadetes, Nitrospirae and candidate division WS3 were more abundant in alkaline soil. We conclude that after 7-years of fertilization, soil bacterial diversity and community structure were shaped more by changes in soil pH rather than the direct effect of nutrient addition.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
The diversity and biogeography of soil bacterial communities.
- Record: found
- Abstract: found
- Article: not found
Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients
- Record: found
- Abstract: found
- Article: not found