- Record: found
- Abstract: found
- Article: found
Tailoring plant-associated microbial inoculants in agriculture: a roadmap for successful application
Read this article at
Abstract
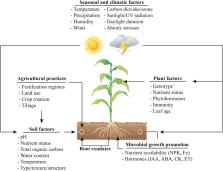
Plants are now recognized as metaorganisms which are composed of a host plant associated with a multitude of microbes that provide the host plant with a variety of essential functions to adapt to the local environment. Recent research showed the remarkable importance and range of microbial partners for enhancing the growth and health of plants. However, plant–microbe holobionts are influenced by many different factors, generating complex interactive systems. In this review, we summarize insights from this emerging field, highlighting the factors that contribute to the recruitment, selection, enrichment, and dynamic interactions of plant-associated microbiota. We then propose a roadmap for synthetic community application with the aim of establishing sustainable agricultural systems that use microbial communities to enhance the productivity and health of plants independently of chemical fertilizers and pesticides. Considering global warming and climate change, we suggest that desert plants can serve as a suitable pool of potentially beneficial microbes to maintain plant growth under abiotic stress conditions. Finally, we propose a framework for advancing the application of microbial inoculants in agriculture.
Abstract
Successful application of plant-associated microbial inoculants for sustainable agriculture requires a holistic approach of examining and understanding the soil environment, crop of interest, and associated microbial communities.
Related collections
Most cited references301
- Record: found
- Abstract: found
- Article: not found
The diversity and biogeography of soil bacterial communities.
- Record: found
- Abstract: found
- Article: not found
The role of root exudates in rhizosphere interactions with plants and other organisms.
- Record: found
- Abstract: found
- Article: not found