- Record: found
- Abstract: found
- Article: found
Overexpressed pseudogenes, DUXAP8 and DUXAP9, promote growth of renal cell carcinoma and serve as unfavorable prognostic biomarkers

Read this article at
Abstract
Background: Growing studies have reported that pseudogenes play key roles in multiple human cancers. However, expression and roles of pseudogenes in renal cell carcinoma remains absent.
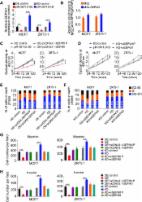
Results: 31 upregulated and 16 downregulated pseudogenes were screened. Higher expression of DUXAP8 and DUXAP9 indicated poorer prognosis of kidney cancer. 33 and 5 miRNAs were predicted to potentially binding to DUXAP8 and DUXAP9, respectively. miR-29c-3p was identified as the most potential binding miRNAs of DUXAP8 and DUXAP9 based on expression, survival and correlation analyses. 254 target genes of miR-29c-3p were forecast. 47 hub genes with node degree >= 10 were identified. Subsequent analysis for the top 10 hub genes demonstrated that COL1A1 and COL1A2 may be two functional targets of DUXAP8 and DUXAP9. Expression of DUXAP8, DUXAP9, COL1A1 and COL1A2 were significantly increased in cancer samples compared to normal controls while miR-29c-3p expression was decreased. Luciferase reporter assay revealed that miR-29c-3p could directly bind to DUXAP8, DUXAP9, COL1A1 and COL1A2. Functional experiments showed that DUXAP8 and DUXAP9 enhanced but miR-29c-3p weakened growth of renal cell carcinoma.
Conclusions: In conclusion, upregulated DUXAP8 and DUXAP9 promote growth of renal cell carcinoma and serve as two promising prognostic biomarkers.
Methods: Dysregulated pseudogenes were obtained by dreamBase and GEPIA. The binding miRNAs of pseudogene and targets of miRNA were predicted using starBase and miRNet. Kaplan-Meier plotter was utilized to perform survival analysis, and Enrichr database was introduced to conduct functional enrichment analysis. Hub genes were identified through STRING and Cytoscape. qRT-PCR, luciferase reporter assay, cell counting assay and colony formation assay were performed to validate in silico analytic results.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression.
- Record: found
- Abstract: found
- Article: found
PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA
- Record: found
- Abstract: found
- Article: not found