- Record: found
- Abstract: found
- Article: not found
Chromatin regulation by Brg1 underlies heart muscle development and disease
Read this article at
SUMMARY
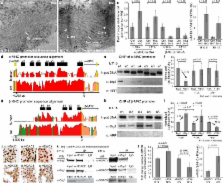
Cardiac hypertrophy and failure are characterized by transcriptional reprogramming of gene expression. Adult cardiomyocytes in mice express primarily α-myosin heavy chain ( α-MHC), whereas embryonic cardiomyocytes express β-MHC. Cardiac stress triggers adult hearts to undergo hypertrophy and a shift from α-MHC to fetal β-MHC expression. Here we show that Brg1, a chromatin-remodeling protein, plays critical roles in regulating cardiac growth, differentiation and gene expression. In embryos, Brg1 promotes myocyte proliferation by maintaining BMP10 and suppressing p57 kip2 expression. It preserves fetal cardiac differentiation by interacting with HDAC and PARP to repress α-MHC and activate β-MHC. In adults, Brg1 is turned off in cardiomyocytes. It is reactivated by cardiac stresses and complexes with its embryonic partners, HDAC and PARP, to induce a pathological α- to β-MHC shift. Preventing Brg1 re-expression decreases hypertrophy and reverses such MHC switch. Brg1 is activated in certain patients with hypertrophic cardiomyopathy, its level correlating with disease severity and MHC changes. Our studies show that Brg1 maintains cardiomyocytes in an embryonic state, and demonstrate an epigenetic mechanism by which three classes of chromatin-modifying factors, Brg1, HDAC and PARP, cooperate to control developmental and pathological gene expression.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Poly(ADP-ribose): novel functions for an old molecule.
- Record: found
- Abstract: found
- Article: not found
Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes.
- Record: found
- Abstract: found
- Article: not found
