- Record: found
- Abstract: found
- Article: found
Resistance patterns among drug-resistant tuberculosis patients and trends-over-time analysis of national surveillance data in Gabon, Central Africa
Read this article at
Abstract
Objective
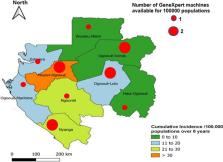
Routinely generated surveillance data are important for monitoring the effectiveness of MDR-TB control strategies. Incidence of rifampicin-resistant tuberculosis (RR-TB) is a key indicator for monitoring MDR-TB.
Methods
In a longitudinal nationwide retrospective study, 8 years (2014–2021) of sputum samples from presumptively drug-resistant tuberculosis patients from all regions of Gabon were referred to the national tuberculosis reference laboratory. Samples were analysed using GeneXpert MTB/RIF and Genotype MTBDRsl version 2/Line Probe Assay.
Results
Of 3057 sputum samples from presumptive tuberculosis patients, both from local hospital and from referral patients, 334 were RR-TB. The median patient age was 33 years (interquartile range 26–43); one third was newly diagnosed drug-resistant tuberculosis patients; one-third was HIV-positive. The proportion of men with RR-TB was significantly higher than that of women (55% vs 45%; p < 0.0001). Patients aged 25–35 years were most affected (32%; 108/334). The cumulative incidence of RR-TB was 17 (95% CI 15–19)/100,000 population over 8 years. The highest incidences were observed in 2020 and 2021. A total of 281 samples were analysed for second-line drug resistance. The proportions of study participants with MDR-TB, pre-XDR-TB and XDR-TB were 90.7% (255/281), 9% (25/281) and 0.3% (1/281), respectively. The most-common mutations in fluoroquinolones resistance isolates was gyrA double mutation gyrA MUT3B and MUT3C (23%; 4/17). Most (64%; 6/8) second-line injectable drugs resistance isolates were characterised by missing both rrs WT2 and MUT2 banding.
Related collections
Most cited references15
- Record: found
- Abstract: found
- Article: not found
The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies.
- Record: found
- Abstract: found
- Article: not found
