- Record: found
- Abstract: found
- Article: not found
Explicit Polarization: A Quantum Mechanical Framework for Developing Next Generation Force Fields
Read this article at
Conspectus

Molecular mechanical force fields have been successfully used to model condensed-phase and biological systems for a half century. By means of careful parametrization, such classical force fields can be used to provide useful interpretations of experimental findings and predictions of certain properties. Yet, there is a need to further improve computational accuracy for the quantitative prediction of biomolecular interactions and to model properties that depend on the wave functions and not just the energy terms. A new strategy called explicit polarization (X-Pol) has been developed to construct the potential energy surface and wave functions for macromolecular and liquid-phase simulations on the basis of quantum mechanics rather than only using quantum mechanical results to fit analytic force fields. In this spirit, this approach is called a quantum mechanical force field (QMFF).
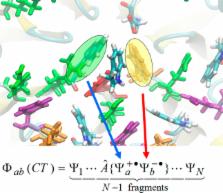
X-Pol is a general fragment method for electronic structure calculations based on the partition of a condensed-phase or macromolecular system into subsystems (“fragments”) to achieve computational efficiency. Here, intrafragment energy and the mutual electronic polarization of interfragment interactions are treated explicitly using quantum mechanics. X-Pol can be used as a general, multilevel electronic structure model for macromolecular systems, and it can also serve as a new-generation force field. As a quantum chemical model, a variational many-body (VMB) expansion approach is used to systematically improve interfragment interactions, including exchange repulsion, charge delocalization, dispersion, and other correlation energies. As a quantum mechanical force field, these energy terms are approximated by empirical functions in the spirit of conventional molecular mechanics. This Account first reviews the formulation of X-Pol, in the full variationally correct version, in the faster embedded version, and with systematic many-body improvements. We discuss illustrative examples involving water clusters (which show the power of two-body corrections), ethylmethylimidazolium acetate ionic liquids (which reveal that the amount of charge transfer between anion and cation is much smaller than what has been assumed in some classical simulations), and a solvated protein in aqueous solution (which shows that the average charge distribution of carbonyl groups along the polypeptide chain depends strongly on their position in the sequence, whereas they are fixed in most classical force fields). The development of QMFFs also offers an opportunity to extend the accuracy of biochemical simulations to areas where classical force fields are often insufficient, especially in the areas of spectroscopy, reactivity, and enzyme catalysis.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Empirical force fields for biological macromolecules: overview and issues.
- Record: found
- Abstract: not found
- Article: not found
Fragmentation methods: a route to accurate calculations on large systems.
- Record: found
- Abstract: found
- Article: not found
