- Record: found
- Abstract: found
- Article: found
Integrating single-cell and bulk RNA sequencing to develop a cancer-associated fibroblast-related signature for immune infiltration prediction and prognosis in lung adenocarcinoma
Read this article at
Abstract
Background
An accumulating amount of studies are highlighting the impacts of cancer-associated fibroblasts (CAFs) on the initiation, metastasis, invasion, and immune evasion of lung cancer. However, it is still unclear how to tailor treatment regimens based on the transcriptomic characteristics of CAFs in the tumor microenvironment of patients with lung cancer.
Methods
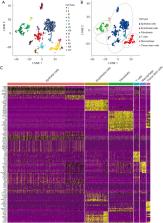
Our study examined single-cell RNA-sequencing data from the Gene Expression Omnibus (GEO) database to identify expression profiles for CAF marker genes and constructed a prognostic signature of lung adenocarcinoma using these genes in The Cancer Genome Atlas (TCGA) database. The signature was validated in 3 independent GEO cohorts. Univariate and multivariate analyses were used to confirm the clinical significance of the signature. Next, multiple differential gene enrichment analysis methods were used to explore the biological pathways related to the signature. Six algorithms were used to assess the relative proportion of infiltrating immune cells, and the relationship between the signature and immunotherapy response of lung adenocarcinoma (LUAD) was explored based on the tumor immune dysfunction and exclusion (TIDE) algorithm.
Results
The signature related to CAFs in this study showed good accuracy and predictive capacity. In all clinical subgroups, the high-risk patients had a poor prognosis. The univariate and multivariate analyses confirmed that the signature was an independent prognostic marker. Moreover, the signature was closely associated with particular biological pathways related to cell cycle, DNA replication, carcinogenesis, and immune response. The 6 algorithms used to assess the relative proportion of infiltrating immune cells indicated that a lower infiltration of immune cells in the tumor microenvironment was associated with high-risk scores. Importantly, we found a negative correlation between TIDE, exclusion score, and risk score.
Related collections
Most cited references83
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found