- Record: found
- Abstract: found
- Article: found
Microenvironmental Conditions Drive the Differential Cyanobacterial Community Composition of Biocrusts from the Sahara Desert
Read this article at
Abstract
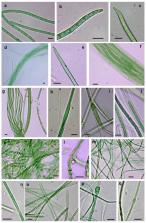
The Sahara Desert is characterized by extreme environmental conditions, which are a unique challenge for life. Cyanobacteria are key players in the colonization of bare soils and form assemblages with other microorganisms in the top millimetres, establishing biological soil crusts (biocrusts) that cover most soil surfaces in deserts, which have important roles in the functioning of drylands. However, knowledge of biocrusts from these extreme environments is limited. Therefore, to study cyanobacterial community composition in biocrusts from the Sahara Desert, we utilized a combination of methodologies in which taxonomic assignation, for next-generation sequencing of soil samples, was based on phylogenetic analysis (16S rRNA gene) in parallel with morphological identification of cyanobacteria in natural samples and isolates from certain locations. Two close locations that differed in microenvironmental conditions were analysed. One was a dry salt lake (a “chott”), and the other was an extension of sandy, slightly saline soil. Differences in cyanobacterial composition between the sites were found, with a clear dominance of Microcoleus spp. in the less saline site, while the chott presented a high abundance of heterocystous cyanobacteria as well as the filamentous non-heterocystous Pseudophormidium sp. and the unicellular cf. Acaryochloris. The cyanobacteria found in our study area, such as Microcoleus steenstrupii, Microcoleus vaginatus, Scytonema hyalinum, Tolypothrix distorta, and Calothrix sp., are also widely distributed in other geographic locations around the world, where the conditions are less severe. Our results, therefore, indicated that some cyanobacteria can cope with polyextreme conditions, as confirmed by bioassays, and can be considered extremotolerant, being able to live in a wide range of conditions.
Related collections
Most cited references101
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.
- Record: found
- Abstract: found
- Article: not found