- Record: found
- Abstract: found
- Article: not found
Modeling Cryptosporidium infection in human small intestinal and lung organoids
Read this article at
Abstract
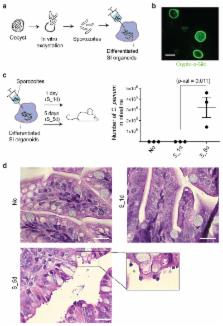
Stem cell-derived organoids recapitulate in vivo physiology of their original tissues, representing valuable systems to model medical disorders such as infectious diseases. Cryptosporidium, a protozoan parasite, is a leading cause of diarrhea and a major cause of child mortality worldwide. Drug development requires detailed knowledge of the pathophysiology of Cryptosporidium, but experimental approaches have been hindered by the lack of an optimal in vitro culture system. Here we show that Cryptosporidium can infect epithelial organoids derived from human small intestine and lung. The parasite propagates within the organoids and completes its complex life cycle. Temporal analysis of the Cryptosporidium transcriptome during organoid infection reveals dynamic regulation of transcripts related to its life cycle. Our study presents organoids as a physiologically relevant in vitro model system to study Cryptosporidium infection.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium.
- Record: found
- Abstract: found
- Article: not found
Cryptosporidium pathogenicity and virulence.
- Record: found
- Abstract: found
- Article: not found
