- Record: found
- Abstract: found
- Article: found
Quantitative Multicolor Super-Resolution Microscopy Reveals Tetherin HIV-1 Interaction
Read this article at
Abstract
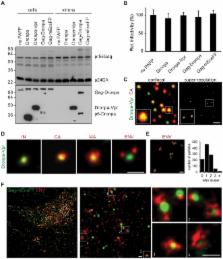
Virus assembly and interaction with host-cell proteins occur at length scales below the diffraction limit of visible light. Novel super-resolution microscopy techniques achieve nanometer resolution of fluorescently labeled molecules. The cellular restriction factor tetherin (also known as CD317, BST-2 or HM1.24) inhibits the release of human immunodeficiency virus 1 (HIV-1) through direct incorporation into viral membranes and is counteracted by the HIV-1 protein Vpu. For super-resolution analysis of HIV-1 and tetherin interactions, we established fluorescence labeling of HIV-1 proteins and tetherin that preserved HIV-1 particle formation and Vpu-dependent restriction, respectively. Multicolor super-resolution microscopy revealed important structural features of individual HIV-1 virions, virus assembly sites and their interaction with tetherin at the plasma membrane. Tetherin localization to micro-domains was dependent on both tetherin membrane anchors. Tetherin clusters containing on average 4 to 7 tetherin dimers were visualized at HIV-1 assembly sites. Combined biochemical and super-resolution analysis revealed that extended tetherin dimers incorporate both N-termini into assembling virus particles and restrict HIV-1 release. Neither tetherin domains nor HIV-1 assembly sites showed enrichment of the raft marker GM1. Together, our super-resolution microscopy analysis of HIV-1 interactions with tetherin provides new insights into the mechanism of tetherin-mediated HIV-1 restriction and paves the way for future studies of virus-host interactions.
Author Summary
Human immunodeficiency virus 1 (HIV-1) assembles and interacts with cellular proteins at the plasma membrane of infected cells. Here, we analyzed individual HIV-1 virions, viral assembly sites and the mechanism of tetherin restriction by multicolor super-resolution microscopy using fully functional fluorescently labeled tetherin and viral proteins. Viral proteins within virions were visualized with nanometer resolution yielding new insight into the structure of the HIV-1. Our super-resolution analysis was extended to tetherin, a cellular restriction factor that inhibits the release of several enveloped viruses. Tetherin was localized in clusters of 70–90 nm at the plasma membrane that contain 5–11 dimers. In contrast tetherin clusters found at HIV-1 assembly sites contained on average 4–7 tetherin dimers. Clustering of tetherin was dependent on both tetherin membrane anchors. The transmembrane domain of tetherin associated with budding virions independently of GM1 lipid raft domains. Our data indicated that extended dimers tether HIV-1 virions directly to the cell. Overall, we provide for the first time super-resolution analysis of authentic virions, virus budding sites and HIV-1 interactions with the anti-viral factor tetherin. Our data offer novel insights into the mechanisms of tetherin restriction.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu.
- Record: found
- Abstract: found
- Article: not found
In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector.
- Record: found
- Abstract: found
- Article: not found