- Record: found
- Abstract: found
- Article: found
pfhrp2 and pfhrp3 Gene Deletions That Affect Malaria Rapid Diagnostic Tests for Plasmodium falciparum: Analysis of Archived Blood Samples From 3 African Countries
Read this article at
Abstract
Background
Malaria rapid diagnostic tests (mRDTs) that target histidine-rich protein 2 (HRP2) are important tools for Plasmodium falciparum diagnosis. Parasites with pfhrp2/3 gene deletions threaten the use of these mRDTs and have been reported in Africa, Asia, and South America. We studied blood samples from 3 African countries to determine if these gene deletions were present.
Methods
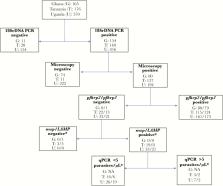
We analyzed 911 dried blood spots from Ghana (n = 165), Tanzania (n = 176), and Uganda (n = 570). Plasmodium falciparum infection was confirmed by 18S rDNA polymerase chain reaction (PCR), and pfhrp2/3 genes were genotyped. True pfhrp2/3 gene deletions were confirmed if samples were (1) microscopy positive; (2) 18S rDNA PCR positive; (3) positive for merozoite surface protein genes by PCR or positive by loop-mediated isothermal amplification; or (4) quantitative PCR positive with >5 parasites/µL.
Abstract
pfhrp2 gene deletions render Plasmodium falciparum parasites undetectable to malaria rapid diagnostic tests detecting histidine-rich protein 2. pfhrp2 deletions were detected in archived blood samples from Tanzania and Uganda, while no samples from Ghana were found to be pfhpr2-negative.
Related collections
Most cited references19
- Record: found
- Abstract: not found
- Article: not found
Nested PCR analysis of Plasmodium parasites.

- Record: found
- Abstract: found
- Article: found
False-negative malaria rapid diagnostic tests in Rwanda: impact of Plasmodium falciparum isolates lacking hrp2 and declining malaria transmission

- Record: found
- Abstract: found
- Article: found