- Record: found
- Abstract: found
- Article: found
APOBEC3G Polymorphism as a Selective Barrier to Cross-Species Transmission and Emergence of Pathogenic SIV and AIDS in a Primate Host
Read this article at
Abstract
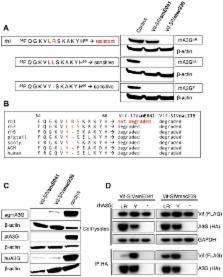
Cellular restriction factors, which render cells intrinsically resistant to viruses, potentially impose genetic barriers to cross-species transmission and emergence of viral pathogens in nature. One such factor is APOBEC3G. To overcome APOBEC3G-mediated restriction, many lentiviruses encode Vif, a protein that targets APOBEC3G for degradation. As with many restriction factor genes, primate APOBEC3G displays strong signatures of positive selection. This is interpreted as evidence that the primate APOBEC3G locus reflects a long-term evolutionary “arms-race” between retroviruses and their primate hosts. Here, we provide direct evidence that APOBEC3G has functioned as a barrier to cross-species transmission, selecting for viral resistance during emergence of the AIDS-causing pathogen SIVmac in captive colonies of Asian macaques in the 1970s. Specifically, we found that rhesus macaques have multiple, functionally distinct APOBEC3G alleles, and that emergence of SIVmac and simian AIDS required adaptation of the virus to evade APOBEC3G-mediated restriction. Our evidence includes the first comparative analysis of APOBEC3G polymorphism and function in both a reservoir and recipient host species (sooty mangabeys and rhesus macaques, respectively), and identification of adaptations unique to Vif proteins of the SIVmac lineage that specifically antagonize rhesus APOBEC3G alleles. By demonstrating that interspecies variation in a known restriction factor selected for viral counter-adaptations in the context of a documented case of cross-species transmission, our results lend strong support to the evolutionary “arms-race” hypothesis. Importantly, our study confirms that APOBEC3G divergence can be a critical determinant of interspecies transmission and emergence of primate lentiviruses, including viruses with the potential to infect and spread in human populations.
Author Summary
APOBEC3G is a host factor that can inhibit replication of primate lentiviruses, including HIV-1, HIV-2, and the related simian immunodeficiency viruses (SIVs) of African primates. As a consequence, primate lentiviruses encode a protein, called Vif, which can induce degradation of APOBEC3G. Given its antiviral role, APOBEC3G may be an important genetic barrier to interspecies jumping of primate lentiviruses. To study this possibility, we asked whether APOBEC3G affected transmission of SIV from sooty mangabeys (SIVsm) to rhesus macaques and subsequent emergence of pathogenic SIVmac in the 1970s. We found that APOBEC3G of sooty mangabeys and rhesus macaques have divergent protein sequences, and that the Vif proteins of SIVsm (Vif-SIVsm) cannot counteract rhesus macaque APOBEC3G. We mapped Vif-SIVsm resistance to a specific substitution in the N-terminal domain of rhesus APOBEC3G, in which a highly conserved tyrosine is replaced by leucine-arginine (Y→LR). We also identified a viral counter-adaptation, found in the Vif proteins of all SIVmac strains, which specifically confers the ability to antagonize APOBEC3G of rhesus macaques. This change was most likely selected during adaptation of SIV to its new host. Together, these results demonstrate that APOBEC3G can serve as a critical genetic determinant of interspecies transmission of primate immunodeficiency viruses.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu.
- Record: found
- Abstract: found
- Article: not found
The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein.
- Record: found
- Abstract: found
- Article: not found