- Record: found
- Abstract: found
- Article: found
Genomic characterization of multidrug‐resistant ESBL‐producing Escherichia coli ST58 causing fatal colibacillosis in critically endangered Brazilian merganser ( Mergus octosetaceus)
Read this article at
Abstract
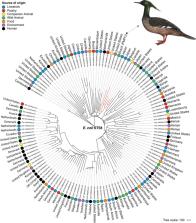
Even though antimicrobial‐resistant bacteria have begun to be detected in wildlife, raising important issues related to their transmission and persistence of clinically important pathogens in the environment, little is known about the role of these bacteria on wildlife health, especially on endangered species. The Brazilian merganser ( Mergus octosetaceus) is one of the most threatened waterfowl in the world, classified as Critically Endangered by the International Union for Conservation of Nature. In 2019, a fatal case of sepsis was diagnosed in an 8‐day‐old Brazilian merganser inhabiting a zoological park. At necropsy, major gross lesions were pulmonary and hepatic congestion. Using microbiologic and genomic methods, we identified a multidrug‐resistant (MDR) extended‐spectrum β‐lactamase (ESBL) CTX‐M‐8‐producing Escherichia coli (designed as PMPU strain) belonging to the international clone ST58, in coelomic cavity, oesophagus, lungs, small intestine and cloaca samples. PMPU strain harboured a broad resistome against antibiotics (cephalosporins, tetracyclines, aminoglycosides, sulphonamides, trimethoprim and quinolones), domestic/hospital disinfectants and heavy metals (arsenic, mercury, lead, copper and silver). Additionally, the virulence of E. coli PMPU strain was confirmed using a wax moth ( Galleria mellonella) infection model, and it was supported by the presence of virulence genes encoding toxins, adherence factors, invasins and iron acquisition systems. Broad resistome and virulome of PMPU contributed to therapeutic failure and death of the animal. In brief, we report for the first time a fatal colibacillosis by MDR ESBL‐producing E. coli in critically endangered Brazilian merganser, highlighting that besides colonization, critical priority pathogens are threatening wildlife. E. coli ST58 clone has been previously reported in humans, food‐producing animals, wildlife and environment, supporting broad adaptation and persistence at human–animal–environment interface.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing.
- Record: found
- Abstract: found
- Article: found