- Record: found
- Abstract: found
- Article: found
Pro-inflammatory megakaryocyte gene expression in murine models of breast cancer

Read this article at
Abstract
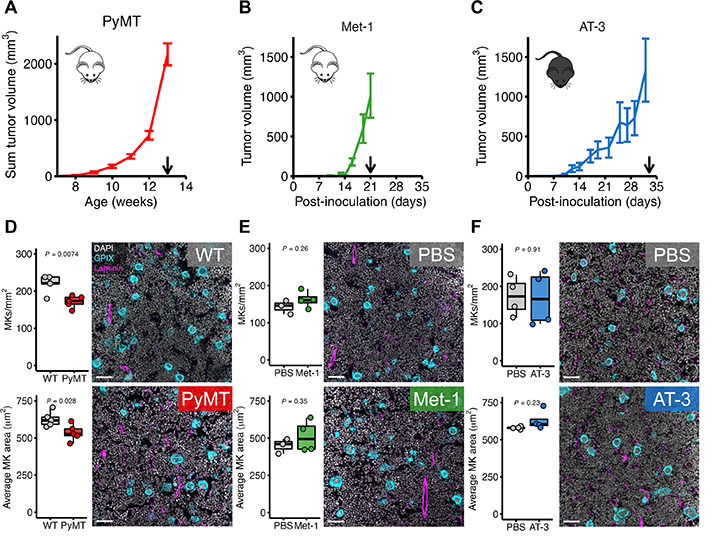
Despite abundant research demonstrating that platelets can promote tumor cell metastasis, whether primary tumors affect platelet-producing megakaryocytes remains understudied. In this study, we used a spontaneous murine model of breast cancer to show that tumor burden reduced megakaryocyte number and size and disrupted polyploidization. Single-cell RNA sequencing demonstrated that megakaryocytes from tumor-bearing mice exhibit a pro-inflammatory phenotype, epitomized by increased Ctsg, Lcn2, S100a8, and S100a9 transcripts. Protein S100A8/A9 and lipocalin-2 levels were also increased in platelets, suggesting that tumor-induced alterations to megakaryocytes are passed on to their platelet progeny, which promoted in vitro tumor cell invasion and tumor cell lung colonization to a greater extent than platelets from wild-type animals. Our study is the first to demonstrate breast cancer–induced alterations in megakaryocytes, leading to qualitative changes in platelet content that may feedback to promote tumor metastasis.
Abstract
Abstract
Breast cancer creates pro-inflammatory megakaryocytes, producing “super-charged” platelets that exacerbate tumor cell metastasis.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Cancer statistics, 2020
- Record: found
- Abstract: found
- Article: not found
Spatial reconstruction of single-cell gene expression
- Record: found
- Abstract: found
- Article: not found