- Record: found
- Abstract: found
- Article: found
Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum
Read this article at
Abstract
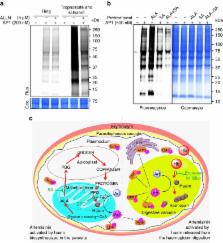
The mechanism of action of artemisinin and its derivatives, the most potent of the anti-malarial drugs, is not completely understood. Here we present an unbiased chemical proteomics analysis to directly explore this mechanism in Plasmodium falciparum. We use an alkyne-tagged artemisinin analogue coupled with biotin to identify 124 artemisinin covalent binding protein targets, many of which are involved in the essential biological processes of the parasite. Such a broad targeting spectrum disrupts the biochemical landscape of the parasite and causes its death. Furthermore, using alkyne-tagged artemisinin coupled with a fluorescent dye to monitor protein binding, we show that haem, rather than free ferrous iron, is predominantly responsible for artemisinin activation. The haem derives primarily from the parasite's haem biosynthesis pathway at the early ring stage and from haemoglobin digestion at the latter stages. Our results support a unifying model to explain the action and specificity of artemisinin in parasite killing.
Abstract
 The mechanism of action of artemisinin, an antimalarial drug, is not well understood.
Here, the authors use a labelled artemisinin analogue to show that the drug is mainly
activated by haem and then binds covalently to over 120 proteins in the malaria parasite,
affecting many of its cellular processes.
The mechanism of action of artemisinin, an antimalarial drug, is not well understood.
Here, the authors use a labelled artemisinin analogue to show that the drug is mainly
activated by haem and then binds covalently to over 120 proteins in the malaria parasite,
affecting many of its cellular processes.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Integration of biological networks and gene expression data using Cytoscape.
- Record: found
- Abstract: found
- Article: not found