- Record: found
- Abstract: found
- Article: found
Partitioning of the denitrification pathway and other nitrite metabolisms within global oxygen deficient zones
Read this article at
Abstract
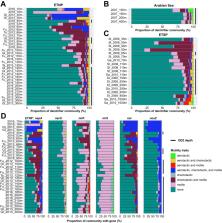
Oxygen deficient zones (ODZs) account for about 30% of total oceanic fixed nitrogen loss via processes including denitrification, a microbially mediated pathway proceeding stepwise from NO 3 – to N 2. This process may be performed entirely by complete denitrifiers capable of all four enzymatic steps, but many organisms possess only partial denitrification pathways, either producing or consuming key intermediates such as the greenhouse gas N 2O. Metagenomics and marker gene surveys have revealed a diversity of denitrification genes within ODZs, but whether these genes co-occur within complete or partial denitrifiers and the identities of denitrifying taxa remain open questions. We assemble genomes from metagenomes spanning the ETNP and Arabian Sea, and map these metagenome-assembled genomes (MAGs) to 56 metagenomes from all three major ODZs to reveal the predominance of partial denitrifiers, particularly single-step denitrifiers. We find niche differentiation among nitrogen-cycling organisms, with communities performing each nitrogen transformation distinct in taxonomic identity and motility traits. Our collection of 962 MAGs presents the largest collection of pelagic ODZ microorganisms and reveals a clearer picture of the nitrogen cycling community within this environment.
Related collections
Most cited references115
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found