- Record: found
- Abstract: found
- Article: not found
Beta- and Novel Delta-Coronaviruses Are Identified from Wild Animals in the Qinghai-Tibetan Plateau, China
Read this article at
Abstract
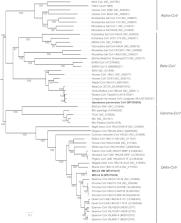
Outbreaks of severe virus infections with the potential to cause global pandemics are increasingly concerning. One type of those commonly emerging and re-emerging pathogens are coronaviruses (SARS-CoV, MERS-CoV and SARS-CoV-2). Wild animals are hosts of different coronaviruses with the potential risk of cross-species transmission. However, little is known about the reservoir and host of coronaviruses in wild animals in Qinghai Province, where has the greatest biodiversity among the world’s high-altitude regions. Here, from the next-generation sequencing data, we obtained a known beta-coronavirus (beta-CoV) genome and a novel delta-coronavirus (delta-CoV) genome from faecal samples of 29 marmots, 50 rats and 25 birds in Yushu Tibetan Autonomous Prefecture, Qinghai Province, China in July 2019. According to the phylogenetic analysis, the beta-CoV shared high nucleotide identity with Coronavirus HKU24. Although the novel delta-CoV (MtCoV) was closely related to Sparrow deltacoronavirus ISU42824, the protein spike of the novel delta-CoV showed highest amino acid identity to Sparrow coronavirus HKU17 (73.1%). Interestingly, our results identified a novel host ( Montifringilla taczanowskii) for the novel delta-CoV and the potential cross-species transmission. The most recent common ancestor (tMRCA) of MtCoVs along with other closest members of the species of Coronavirus HKU15 was estimated to be 289 years ago. Thus, this study increases our understanding of the genetic diversity of beta-CoVs and delta-CoVs, and also provides a new perspective of the coronavirus hosts.
Related collections
Most cited references37

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
