- Record: found
- Abstract: found
- Article: found
Degradation reduces the diversity of nitrogen-fixing bacteria in the alpine wetland on the Qinghai-Tibet Plateau
Read this article at
Abstract
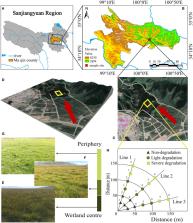
Biological nitrogen fixation is a key process in the nitrogen cycle and the main source of soil available nitrogen. The number and diversity of nitrogen-fixing bacteria directly reflect the efficiency of soil nitrogen fixation. The alpine wetland on the Qinghai-Tibet Plateau (QTP) is degrading increasingly, with a succession toward alpine meadows. Significant changes in soil physicochemical properties accompany this process. However, it is unclear how does the soil nitrogen-fixing bacteria change during the degradation processes, and what is the relationship between these changes and soil physicochemical properties. In this study, the nifH gene was used as a molecular marker to further investigate the diversity of nitrogen-fixing bacteria at different stages of degradation (none, light, and severe degeneration) in the alpine wetland. The results showed that wetland degradation significantly reduced the diversity, altered the community composition of nitrogen-fixing bacteria, decreased the relative abundance of Proteobacteria, and increased the relative abundance of Actinobacteria. In addition to the dominant phylum, the class, order, family, and genus of nitrogen-fixing bacteria had significant changes in relative abundance. Analysis of Mantel test showed that most soil factors (such as pH, soil water content (SWC), the organic carbon (TOC), total nitrogen (TN), and soil C:P ratio) and abundance had a significant positive correlation. TOC, TN, total phosphorus (TP), soil C:P ratio and Shannon had a significant positive correlation with each other. The RDA ranking further revealed that TOC, SWC, and TN were the main environmental factors influencing the community composition of nitrogen-fixing bacteria. It is found that the degradation of the alpine wetland inhibited the growth of nitrogen-fixing bacteria to a certain extent, leading to the decline of their nitrogen-fixing function.
Related collections
Most cited references60

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: found
- Article: not found
UPARSE: highly accurate OTU sequences from microbial amplicon reads.
- Record: found
- Abstract: found
- Article: not found