- Record: found
- Abstract: found
- Article: found
Identification of gene signatures associated with ulcerative colitis and the association with immune infiltrates in colon cancer
Read this article at
Abstract
Background
Inflammatory bowel diseases, including ulcerative colitis (UC) and Crohn’s disease, are some of the most common inflammatory disorders of the gastrointestinal tract. The dysfunction of the immune system in the intestines is suggested to be the underlying cause of the pathogenesis of UC. However, the mechanisms regulating these dysfunctional immune cells and inflammatory phenotypes are still unclear.
Methods
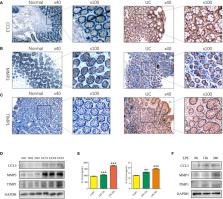
The differential expression analysis on microarray datasets were performed including GSE24287, GSE87466, GSE102133, and GSE107499, including 376 samples. “Gene Ontology” and “Kyoto Encyclopedia of Genes and Genomes” pathway enrichment analyses were conducted to identify the common differentially expressed genes (DEGs) in these datasets and explore their underlying biological mechanisms. Further algorithms like “Cell-type Identification by Estimating Relative Subsets of RNA Transcripts” were used to determine the infiltration status of immune cells in patients with UC. “Cytoscape” and “Gene Set Enrichment Analysis” were used to screen for hub genes and to investigate their biological mechanisms. The Tumor Immune Estimation Resource database was used to study the correlation between hub genes and infiltrating immune cells in patients with UC. A total of three hub genes, CCL3, MMP3, and TIMP1, were identified using Cytoscape.
Results
A positive correlation was observed between these hub genes and patients with active UC. These genes served as a biomarker for active UC. Moreover, a decrease in CCL3, MMP3, and TIMP1 expression was observed in the mucosa of the intestine of patients with active UC who responded to Golimumab therapy. In addition, results show a significant positive correlation between CCL3, MMP3, and TIMP1 expression and different immune cell types including dendritic cells, macrophages, CD8+ T cells, and neutrophils in patients with colon cancer. Moreover, CCL3, MMP3, and TIMP1 expression were strongly correlated with different immune cell markers.
Conclusion
Study results show the involvement of hub genes like CCL3, MMP3, and TIMP1 in the pathogenesis of UC. These genes could serve as a novel pharmacological regulator of UC. These could be used as a therapeutic target for treating patients with UC and may serve as biomarkers for immune cell infiltration in colon cancer.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: found
Hallmarks of Cancer: The Next Generation

- Record: found
- Abstract: found
- Article: found
Metascape provides a biologist-oriented resource for the analysis of systems-level datasets

- Record: found
- Abstract: found
- Article: found