- Record: found
- Abstract: found
- Article: found
Alterations in the gut virome in patients with ankylosing spondylitis
Read this article at
Abstract
Introduction
Ankylosing spondylitis (AS), a chronic autoimmune disease, has been linked to the gut bacteriome.
Methods
To investigate the characteristics of the gut virome in AS, we profiled the gut viral community of 193 AS patients and 59 healthy subjects based on a metagenome-wide analysis of fecal metagenomes from two publicly available datasets.
Results
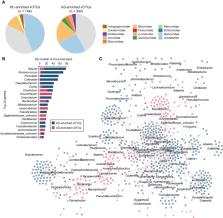
AS patients revealed a significant decrease in gut viral richness and a considerable alteration of the overall viral structure. At the family level, AS patients had an increased abundance of Gratiaviridae and Quimbyviridae and a decreased abundance of Drexlerviridae and Schitoviridae. We identified 1,004 differentially abundant viral operational taxonomic units (vOTUs) between patients and controls, including a higher proportion of AS-enriched Myoviridae viruses and control-enriched Siphoviridae viruses. Moreover, the AS-enriched vOTUs were more likely to infect bacteria such as Flavonifractor, Achromobacter, and Eggerthellaceae, whereas the control-enriched vOTUs were more likely to be Blautia, Ruminococcus, Collinsella, Prevotella, and Faecalibacterium bacteriophages. Additionally, some viral functional orthologs differed significantly in frequency between the AS-enriched and control-enriched vOTUs, suggesting the functional role of these AS-associated viruses. Moreover, we trained classification models based on gut viral signatures to discriminate AS patients from healthy controls, with an optimal area under the receiver operator characteristic curve (AUC) up to 0.936, suggesting the clinical potential of the gut virome for diagnosing AS.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found
fastp: an ultra-fast all-in-one FASTQ preprocessor
- Record: found
- Abstract: found
- Article: not found