- Record: found
- Abstract: found
- Article: found
Causal associations between insulin-like growth factor 1 and vitamin D levels: a two-sample bidirectional Mendelian randomization study
Read this article at
Abstract
Background
Insulin-like growth factor 1 (IGF-1) plays a vital role in the attainment and maintenance of bone mass throughout life and is closely related to the stature of children. 25-Hydroxyvitamin D (25-OHD) is an intermediate of vitamin D (Vit D) metabolism and a key indicator of Vit D nutritional status. Multiple studies have revealed that IGF-1 levels undergo a non-significant increase after Vit D supplementation. Here, we analyzed the causal and reverse causal relationships between 25-OHD and IGF-1 levels using Mendelian randomization (MR).
Methods
Two-sample MR was used to estimate an unconfounded bidirectional causal relationship between the level of IGF-1 and those of Vit D and 25-OHD. Single nucleotide polymorphisms (SNPs) were filtered from genome-wide association studies (GWAS) after a comprehensive search of the Integrative Epidemiology Unit GWAS database. Several MR methods were employed, including inverse-variance weighted (IVW) method, and a sensitivity analysis was undertaken to detect whether pleiotropy or heterogeneity biased the MR results.
Results
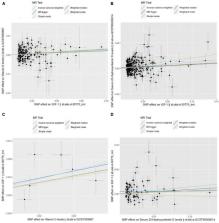
Genetically predicted IGF-1 was found to have a causal association with Vit D and serum 25-OHD levels, where Vit D and serum 25-OHD levels increased with increasing IGF-1 concentrations (Vit D: IVW β:0.021, 95% CI: 0.005–0.036, p = 7.74 × 10 –3; 25-OHD: IVW β: 0.041, 95% CI: 0.026–0.057, p = 2.50 × 10 –7). A reverse causal effect was also found, indicating Vit D and serum 25-OHD have a positive causal relationship with IGF-1 (Vit D: IVW β:0.182, 95% CI: 0.061–0.305, p = 3.25 × 10 –3; 25-OHD: IVW β: 0.057, 95% CI = 0.017–0.096, p = 4.73 × 10 –3). The sensitivity analysis showed that horizontal pleiotropy was unlikely to bias the causality in this study (MR-Egger: Vit D intercept p = 5.1 × 10 –5, 25-OHD intercept p = 6.4 × 10 –4 in forward analysis; Vit D intercept p = 6.6 × 10 –4, 25-OHD intercept p = 1.9 × 10 –3 in reverse analysis), and a leave-one-out analysis did not identify evidence of bias in the results.
Conclusion
The results of the MR analysis provide evidence that IGF-1 has positive causal and reverse causal relationships with Vit D and serum 25-OHD, respectively, in European populations. Our findings also provide guidance for the prevention and treatment of short stature and other related diseases.
Related collections
Most cited references47

- Record: found
- Abstract: found
- Article: found
Second-generation PLINK: rising to the challenge of larger and richer datasets

- Record: found
- Abstract: found
- Article: found
Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression

- Record: found
- Abstract: found
- Article: found